
For a full list of BASHing data blog posts see the index page. ![]()
Data quality in iNaturalist downloads
iNaturalist is a citizen science platform. Take a picture of something (a flower, a bird, a frog) and upload it to iNaturalist. If the image is properly dated and georeferenced, and if the organism is free in nature (not captive or cultivated), then your observation is "verifiable". Verifiables with a solid identification achieve "research grade" status. These are observations for which two-thirds of the experts in the iNaturalist community agree on an ID.
Research-grade observations document the presence of a particular species on a particular day at a particular place. They're passed on to GBIF, the world's largest aggregator of biological occurrence records. GBIF processes the records further, sometimes disagreeing with the name or classification of the organism given in iNaturalist. (See below for an example.)
All records are available on the iNaturalist website as webpages with images. Records can also be downloaded, although currently there's a download limit of 200,000 records per batch.
I've been impressed with the quality of iNaturalist data, both in iNaturalist downloads and in GBIF downloads. To be clear, I don't mean "quality" in the scientific sense of accuracy or credibility. I mean that iNaturalist's plain-text records are largely free of the many mistakes and problems detailed in A Data Cleaner's Cookbook. iNaturalist records aren't perfect, but they require very little cleaning.
For example, I recently downloaded 34,718 iNaturalist records for arachnids in Australia and I found only a small number of structure and content errors; these were all in fields into which users can freely enter text. The only format problem I found was Windows line endings in 1025 of the records.
Checking characters in iNaturalist records is a challenge because users enter not only numbers and Latin alphabet characters, but also non-Western characters (like 澳 , U+6FB3), emojis, text emoticons (":-)") and picture glyphs (🕷, U+1F577). My records sample, however, had only a handful of gremlins (nine in total, in eight records) in the free-text "description" field. The download's encoding was UTF-8.
Most of the gremlins were reconstructable replos. In one record the data-enterer had put an "en dash" between page numbers instead of a hyphen. The UTF-8 encoding for an "en dash" is hexadecimal e2 80 93. This was evidently first processed byte-by-byte and the three bytes were interpreted as a Latin small letter "a" with circumflex (â) and the undefined characters encoded as 80 and 93. Back to UTF-8, and the three characters became "â" (hex c3 a2), the "padding" control character (c2 80) and the "set transmit state" control character (c2 93).
Six records mentioned Uluru-Kata Tjuta National Park, but spelled the Aboriginal words with modern orthography: Uluṟu-Kata Tjuṯa. The "ṟ" has hex value e1 b9 9f in UTF-8, and the "ṯ" is e1 b9 af. Once again there seems to have been byte-by-byte processing followed by a return to UTF-8. The result was Uluá¹APCu-Kata Tjuṯa, where "á" is c3 a1 (e1 in Windows-1252), "¹" is c2 b9, the "application program command" control character APC is c2 9f, and the macron ¯ is c2 af.
From a data cleaner's point of view, a bigger problem with iNaturalist downloads is that they're only available in the problematic CSV format, and many records have embedded newlines. I wasn't able to cleanly convert my (tab-free) download "observations-76494.csv" to a TSV for auditing using either csvformat from the csvkit bundle of utilities
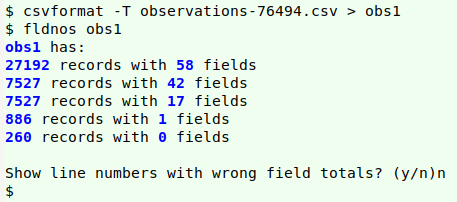
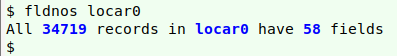
or the c2t function from the Cookbook. My sample dataset was only 20 MB, luckily, so I was able to open it as a spreadsheet in LibreOffice Calc, then copy and paste the active cells as a tab-separated text file, "locar0". Calc converts each embedded newline to a single whitespace in this process.
The embedded newline issue is also fixed in GBIF and in the Atlas of Living Australia (ALA), which aggregates Australian iNaturalist records. For example, this iNaturalist record has the "description" field contents spread over five lines, two of which are blank:

In the corresponding GBIF record (here; accessed 2020-01-27) the "description" entry is filed under "occurrenceRemarks", the blank lines are gone and the strings are separated by a single whitespace. In ALA (record here; accessed 2020-01-27) the blank lines are gone but there's no spacing between strings. (In the LibreOffice Calc conversion, two newlines and a blank line in the cell were replaced with two whitespaces during copy/pasting.)
Note that iNaturalist calls this spider Trichonephila plumipes. It gets that name from an authoritative source, the World Spider Catalog, here. The World Spider Catalog classification is in turn based on a 2019 scientific paper in which the species Nephila plumipes is moved to the genus Trichonephila based on molecular-genetic evidence.
ALA isn't as up-to-date and follows the Australian Faunal Directory (AFD) for scientific names. The AFD says that Trichonephila is a synonym of Nephila, and ALA "up-matches" the iNaturalist record to Nephila. However, the AFD recognises the species name plumipes and calls this spider Nephila plumipes, so it's not clear why ALA didn't call it that as well.
GBIF follows yet another authority, its own "backbone taxonomy". Trichonephila plumipes isn't listed there, and GBIF replaces it with the genus name Trichonephila, which it accepts.
So Trichonephila plumipes for this one record in iNaturalist, Nephila in ALA and Trichonephila in GBIF. If you find these disagreements between record-aggregating projects a little confusing, you're not alone. So do biologists. Unfortunately, scientific names aren't the only things that record-aggregators tinker with when processing original data. (Don't get me started.) And data quality in GBIF and ALA records from museums and herbaria can be pretty awful.
Which is why the up-to-date and relatively non-messy records in iNaturalist impress me. Now, if they could just fix that CSV/embedded newline issue...
Last update: 2020-02-05
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License