TSV This marker means that the recipe only works with tab-separated data tables.
Fix Windows line endings
On Linux and Apple computers, a newline is built with just one character, the UNIX linefeed "\n" (LF, hex 0a). On Windows computers, a newline is created using two characters, one after the other: "\r\n", where "\r" is called a "carriage return" (CR, hex 0d). CRLFs aren't necessary in a data table and can cause serious problems in data cleaning. For this reason, CRLF line endings should be converted to LF line endings before data checking and cleaning is done.
Detect and count CRs, in total and at line endings
CRcheck() { grep -oP "\r" "$1" | wc -l && grep -oP "\r$" "$1" | wc -l; }
The "CRcheck" function generates a count of all CRs, then a count of CRs in CRLF line endings. If the two counts are the same, then all the CRs in table are part of CRLF line endings, and the CRs can be removed globally:
Delete all CRs
tr -d "\r" < table > table_noCRs
If the first count is larger than the second count, then there are one or more CRs somewhere in table that are not part of CRLF line endings. An easy way to find them is to first delete the CRs in CRLF line endings:
Delete CRs in line endings only
sed 's/\r$//' table > table_noCRLFs
Next, find the records with internal CRs and their record numbers.
Locate CRs not in line endings
cat -v table_noCRLFs | grep -n "\^M"
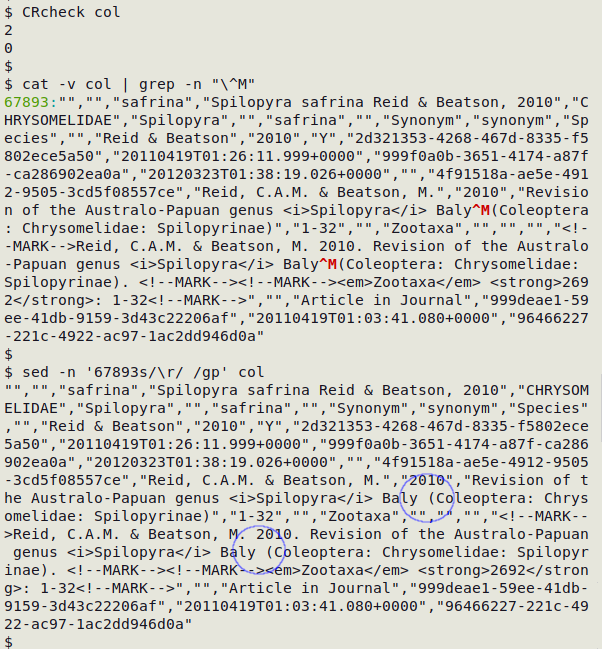
Finally, decide how to get rid of the internal CRs. In the real-world case shown below, I used sed to replace each of the two internal CRs in line 67893 of the CSV "col" with a single whitespace.
Multiple character versions
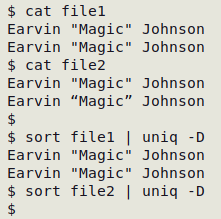
It's good, tidy practice to have just one version of each character in a data table. If more than one version is used, duplicates may be missed. In the example shown below, "file2" has left and right double quotes in the second line instead of plain double quotes:
Multiple versions of single characters are easily detected with the "graph" script, which also displays UTF-8 encodings in hexadecimal. Replace more complicated versions with simple ones:

Unmatched braces
The second example used to illustrate the "graph" script shows that the table "ver1" has 143728 "(" but only 143726 ")", and 10122 "[" but only 10119 "]". The unmatched braces might just be typographical errors, or they might indicate that a closing brace was lost when a data item was truncated.
The "unmatched" function (below) locates unmatched braces within tab-separated fields. It returns the line (record) number and field number, then the whole data item in the field. I use it for the pairs (), [] and {}. Shown here is the beginning of the result for round braces in "ver1":

Locate data items containing unmatched braces TSV
(The second and third arguments are the opening and closing braces, in quotes)
unmatched() { awk -F"\t" -v start="$2" -v end="$3" '{for (i=1;i<=NF;i++) if (split($i,a,start) != split($i,b,end)) print "line "NR", field "i":\n"$i}' "$1"; }
"graph" and "unmatched" will not find data items with matched but backwards braces, as in aaa)aaa(aaa. Cases like these could be found with a regular expression:
awk -F"\t" '{for (i=1;i<=NF;i++) if ($i ~ /^[^()]*\)[^()]*\(/) {print "line "NR", field "i": "$i}}' table
A much faster and more informative way to locate unmatched braces in a TSV is to use one of the following three functions, which look respectively for unmatched parentheses ("umparID"), unmatched square braces ("umsquID") and unmatched curly braces ("umcurID"). Each function takes as arguments the TSV filename and the field number of a field which has a unique ID code for each record. The functions return a pipe-separated list with the unique ID code, the name of the field with the unmatched braces, and the data item in that field. The braces are highlighted with a yellow background, and the list is sorted first by field name and second by unique ID code.
umparID() { echo "ID | field name | data item"; awk -F"\t" -v idfld="$2" 'FNR==NR {if (NR==1) for (i=1;i<=NF;i++) a[i]=$i; else if (gsub(/\(/,"") != gsub(/\)/,"")) b[NR]; next} FNR in b {for (j=1;j<=NF;j++) if (split($j,c,"(") != split($j,d,")")) print $idfld FS a[j] FS $j}' "$1" "$1" | sort -t $'\t' -k2,2 -Vk1,1 | sed $'s/\t/ | /g;s/(/\033[103m(\033[0m/g;s/)/\033[103m)\033[0m/g'; }
umsquID() { echo "ID | field name | data item"; awk -F"\t" -v idfld="$2" 'FNR==NR {if (NR==1) for (i=1;i<=NF;i++) a[i]=$i; else if (gsub(/\[/,"") != gsub(/\]/,"")) b[NR]; next} FNR in b {for (j=1;j<=NF;j++) if (split($j,c,"[") != split($j,d,"]")) print $idfld FS a[j] FS $j}' "$1" "$1" | sort -t $'\t' -k2,2 -Vk1,1 | sed $'s/\t/ | /g;s/[[]/\033[103m&\033[0m/g;s/[]]/\033[103m&\033[0m/g'; }
umcurID() { echo "ID | field name | data item"; awk -F"\t" -v idfld="$2" 'FNR==NR {if (NR==1) for (i=1;i<=NF;i++) a[i]=$i; else if (gsub(/\{/,"") != gsub(/\}/,"")) b[NR]; next} FNR in b {for (j=1;j<=NF;j++) if (split($j,c,"{") != split($j,d,"}")) print $idfld FS a[j] FS $j}' "$1" "$1" | sort -t $'\t' -k2,2 -Vk1,1 | sed $'s/\t/ | /g;s/[{]/\033[103m&\033[0m/g;s/[}]/\033[103m&\033[0m/g'; }
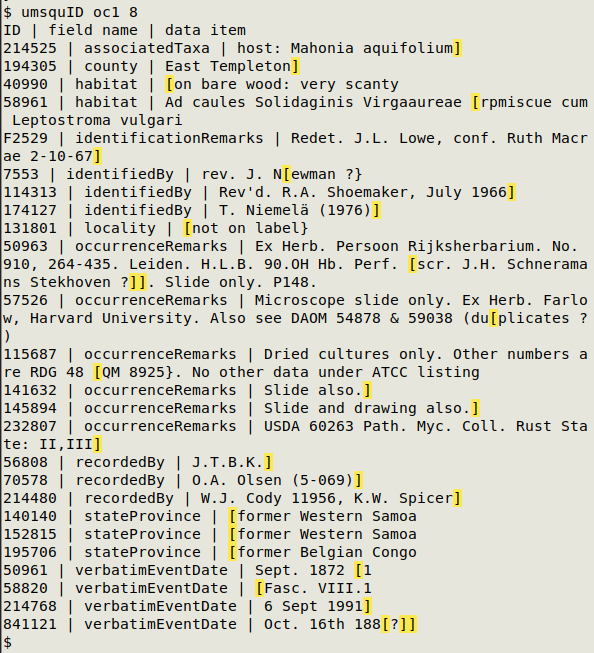
In the screenshot below, "umsquID" is finding unmatched square braces in the file oc1, with field 8 used for a unique ID.
Excess whitespace
As with multiple character versions, excess whitespace can create pseudo-duplicates and mask duplication:
Watch this space
Watch this space
Watch this space
Watch this space
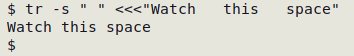
Trim any/all series of whitespace characters to one whitespace each
tr -s " " < table > trimmed_table
Locate leading or trailing whitespace within data items TSV
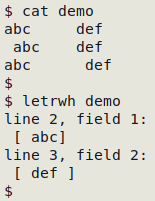
letrwh() { awk -F"\t" '{for (i=1;i<=NF;i++) {if ($i ~ /^[ ]+/ || $i ~ /[ ]+$/) print "line "NR", field "i":\n ["$i"]"}}' "$1"; }
The "letrwh" function detects leading or trailing whitespace within a data item and prints the line (record) number and the field number, then the data item with enclosing square braces, so the leading or trailing whitespace can be more easily seen:
Delete all leading or trailing whitespace within data items TSV
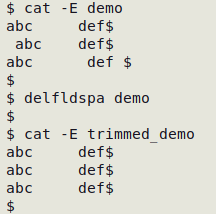
delfldspa() { awk 'BEGIN{FS=OFS="\t"} {for (i=1;i<=NF;i++) gsub(/^[ ]+|[ ]+$/,"",$i); print}' "$1" > "trimmed_$1"; }
The "delfldspa" function removes leading and trailing whitespace in all data items and builds a new, trimmed data table
Malformed markup
When auditing data tables I sometimes find malformed HTML markup. The problem is nicely illustrated by this real-world example:

The tags <i> and </i> were originally used to put an enclosed string in italics, maybe on a web page or in a program that understood HTML markup. The markup is correct around Brotia but is malformed around Zoosystematics and Evolution. In addition to malformed HTML tags, I sometimes see correctly formed tags in the wrong place, such as in </i>some text<i>, as well as non-HTML markup, such as <italic>, <bold> and <roman>.
How to find malformed and misplaced markup efficiently on the command line? Checking every instance of < and > would be painfully slow, and I haven't yet found any validator or parser for HTML that can identify all these markup problems, mainly because I'm not actually auditing HTML documents.
My solution isn't very efficient, but it seems to work. The method has two stages and uses two small, reference files — "openers" and "closers" — listing the HTML tags I'm most likely to find in data tables. The files are newline-separated lists but are printed below as comma-separated files to save space:
The actual files are newline-separated lists!
"openers"
<b>,<B>,<em>,<EM>,<h1>,<H1>,<h2>,<H2>,<h3>,<H3>,<h4>,<H4>,<h5>,<H5>,<h6>,<H6>,<i>,<I>,<p>,<P>,<strong>,<STRONG>,<sub>,<SUB>,<sup>,<SUP>
"closers"
</b>,</B>,<br>,<br />,<BR>,<BR />,</em>,</EM>,</h1>,</H1>,</h2>,</H2>,</h3>,</H3>,</h4>,</H4>,</h5>,</H5>,</h6>,</H6>,</i>,</I>,</p>,</P>,</strong>,</STRONG>,</sub>,</SUB>,</sup>,</SUP>
The first stage uses a while loop to read one of the reference files, then searches with grep for occurrences of tags that might be "improperly" adjoining alphanumeric characters, or isolated by spaces on either side:
while read line; do grep -E "[[:alnum:]]$line|[[:blank:]]$line[[:blank:]]" table; done < path/to/openers
while read line; do grep -E "$line[[:alnum:]]|[[:blank:]]$line[[:blank:]]" table; done < path/to/closers
This first stage also finds awkward but valid placements of tags, such as <em>string[space]</em> instead of <em>string</em>[space].
The second stage command first replaces all the correctly formed tags with "{MARKUP}" and prints a record number for each record, then looks for stray < and > in the modified data table:
awk 'FNR==NR {a[$0]=$0; next} {for (i in a) gsub(a[i],"{MARKUP}"); print FNR": "$0}' <(cat path/to/openers path/to/closers) table | grep -E "<|>"
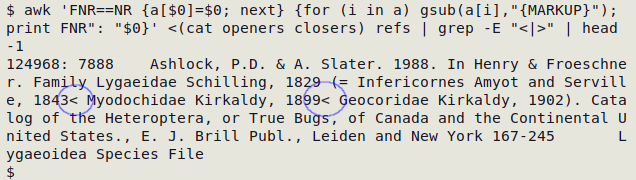
The screenshot below shows the first result of the second-stage command at work on the table "refs". The command has found two stray < in "refs".