
For a full list of BASHing data blog posts see the index page. ![]()
Spellchecking scientific names on the command line
Writing a spellchecking program seems to be a popular coding exercise. When I searched GitHub for "spellchecker" last month I found 1,643 projects. So far as I know, most of the programs work the same way. Given a word, the spellchecker looks to see if that word is in a reference list of correctly spelled words. If there's a successful match, then the word is correctly spelled. If not, the word is misspelled.
The widely used and more sophisticated spellchecker hunspell works differently, and tries to find a match by combining a word root with a prefix or suffix, or transforming a word in a language-compliant way. Clever programs like aspell will also suggest alternatives to misspellings and allow you to replace the misspelled word with a selected alternative.
I wanted a basic spellchecker for the scientific names I see in my data auditing work, so I generated a reference list of names (see below) and then devised a couple of simple functions to do the checking. The functions are based on AWK, and to show how they work I'll use the /usr/share/dict/american-english on my system as a reference list. It's a plain-text file with 102,041 words, one word per line. The sample to be spellchecked is the file "to_check":
sieze
exceed
accomodate
independant
maintenance
If I put the whole reference list in an AWK array as index strings, I can then see which items in "to_check" are not index strings in the array:
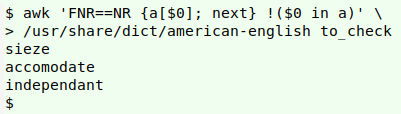
awk 'FNR==NR {a[$0]; next} !($0 in a)' \
/usr/share/dict/american-english to_check
A custom dictionary. My source for scientific names is the GBIF Backbone Taxonomy (GBT). Although the GBT is far from complete and contains formatting and other errors, it's a reasonably comprehensive compilation of the scientific names of animals, plants, fungi, bacteria etc.
I downloaded the latest GBT archive on 2020-04-06. From the archive I extracted the "Taxon.tsv" table. Field 8 in that table is "canonicalName" and contains scientific names stripped of author-date strings (Eopenthes deceptor Sharp, 1908 Eopenthes deceptor) and free of problems like formatting for hybrids (×Thompsoveria Thompsoveria) and disallowed characters (Blötea Blotea).
The entries in field 8 can contain 1, 2 or 3 words, with multiple words separated by a single space, e.g. Sanguinolitidae, Baputa dichroa and Crepidotus variabilis subsphaerosporus. To build a reference list with one component on each line I cut out field 8 below the header, removed any blank lines with AWK, converted the spaces between words to newlines with tr, then sorted and uniquified the output and saved the resulting list in "gbtnames":
tail -n +2 Taxon.tsv | cut -f8 | awk NF | tr " " "\n" | sort | uniq > gbtnames
I then deleted 105 erroneous entries from "gbtnames", leaving a wordlist of 1,309,645 scientific name components as my custom dictionary.
Most of the errors in "canonicalName" are proofreading failures. For example, after harvesting the scientific name "Oberea near strigosa Shelford, 1902", GBIF parsed that as genus "Oberea", species "near", subspecies "strigosa". But the specialist who wrote the name string was saying "This is an Oberea species close to strigosa Shelford, 1902".
Some of the checking failures are pretty obvious (there really isn't a species called Unidentified unidentified Hood [accessed 2020-05-05]) but others are more subtle and I may have missed some. Note that "105" is only the number of unique strings I deleted from my "gbtnames"; the number of names containing these strings in GBT's "Taxon.tsv" is much larger.
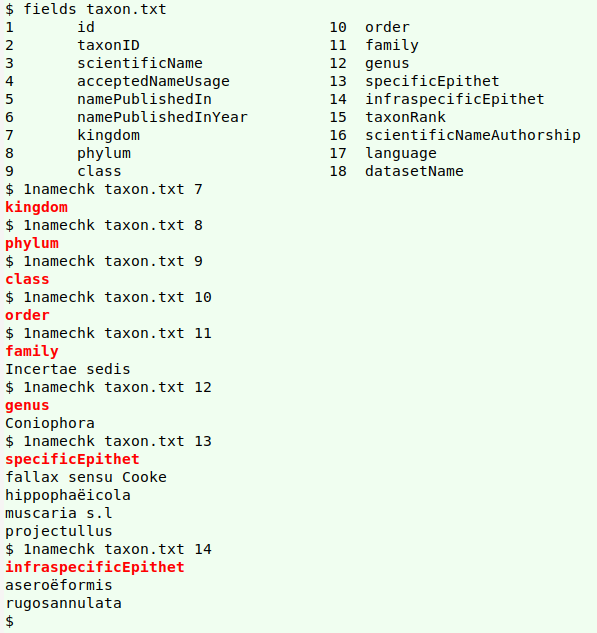
Checking real-world data. Most of my name-checking is in tab-separated tables where particular fields contain scientific names. My first checker is called "1namechk", and it's for fields containing one-word names. I give "1namechk" the name of the TSV to be checked and the number of the field to be checked, and AWK returns names in that field that are not in "gbtnames", which lives in my "scripts" folder. The AWK command also ignores any blank entries in the field. The possibly misspelled names are then sorted and uniquified.
I haven't excluded the header line, so that a field with no potential misspellings returns the field name instead of nothing at all. However, for clarity I've made the field name red and bold in the results with sub.
In my locale, GNU sort puts strings beginning with the escape character ("\033"; hex 1b) between strings beginning with punctuation and strings beginning with alphanumeric characters. Try this command in your locale:
printf "bbb\n,,,\n\033[1;31mword\033[0m\n333\n" | sort
1namechk() { awk -v fld="$2" -F"\t" 'FNR==NR {a[$0]; next} FNR==1 {sub($fld,"\033[1;31m&\033[0m")} $fld != "" && !($fld in a) {print $fld}' ~/scripts/gbtnames "$1" | sort | uniq; }
Here are the results of successive one-name checks on fields 7-14 in the real-world table "taxon.txt":
A more elegant way to do this on a range of fields is with the following command, which only builds the big array once, rather than each time a field is examined:
awk -F"\t" 'FNR==NR {a[$0]; next} {for (i=7;i<=14;i++) {if (FNR==1) sub($i,"\033[1m&\033[0m"); if ($i != "" && !($i in a)) print i FS $i}}' ~/scripts/gbtnames taxon.txt | sort -n | uniq
The corresponding function "1namechkR" takes three arguments:
1namechkR [filename] [start field] [end field]:
1namechkR() { awk -v start="$2" -v end="$3" -F"\t" 'FNR==NR {a[$0]; next} {for (i=start;i<=end;i++) {if (FNR==1) sub($i,"\033[1;31m&\033[0m"); if ($i != "" && !($i in a)) print i FS $i}}' ~/scripts/gbtnames "$1" | sort -n | uniq; }
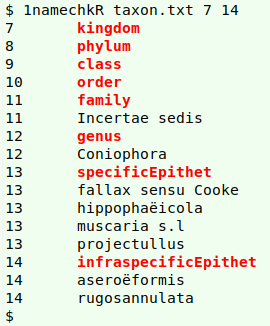
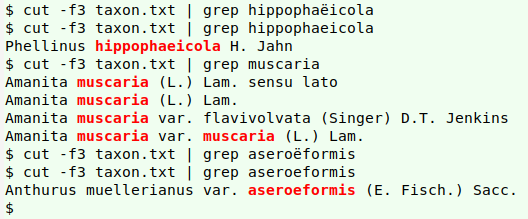
As shown in the screenshot above, there are problems in the "genus" (12), "specificEpithet" (13) and "infraspecificEpithet" (14) fields. The "Incertae sedis" in the "family" field (11) is OK. It's not a name, but a Latin phrase indicating (in this case) that the species has not yet been assigned to a family.
A grep search in the GBT's "Taxon.tsv" turns up a number of species in the genus Coniophora, so why no match for that genus? Because Coniophora in the real-world table ends with an invisible no-break space (hex c2 a0), a gremlin character:
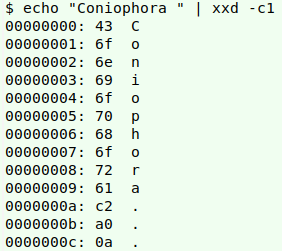
Memo to self: clean up gremlins and leading/trailing spaces before doing name checks.
Next, the two names with diacritical marks over an "e" are indeed in "gbtnames", but without the marks:

"specificEpithet" and "infraspecificEpithet" have some other problems. In "specificEpithet", the strings "fallax sensu Cooke" and "muscaria s.l" are invalid entries. The field "specificEpithet" should only contain the Latin species name, and any comments on the name should be in a different field.
"fallax sensu Cooke" means the name "fallax" as understood by Cooke. "muscaria s.l" (should be "muscaria s.l."), for "muscaria sensu lato", means "muscaria" in the broad sense; this includes the species "muscaria" and any subspecies, as well as species that have been called "muscaria" in the past, although they may not, in fact, belong in "muscaria".
That leaves a couple of possibly misspelled names found by the spellcheck: "projectullus" and "rugosannulata":
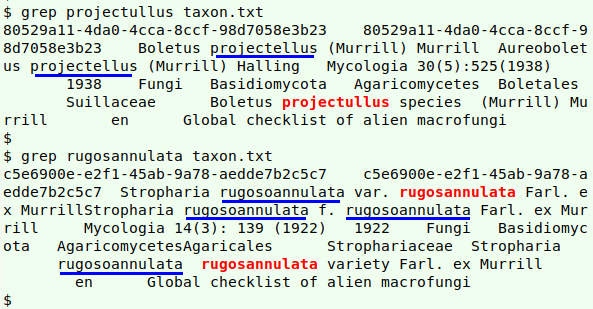
grepping them in the file being checked (screenshot above) suggests that "projectullus" (in red) is a misspelling of "projectellus" (blue underlines), and a little googling shows that to be true. (The species is now called Aureoboletus projectellus.) Similarly, "rugosannulata" looks like a misspelling of "rugosoannulata", and it's indeed a misspelling, according to the GBT:
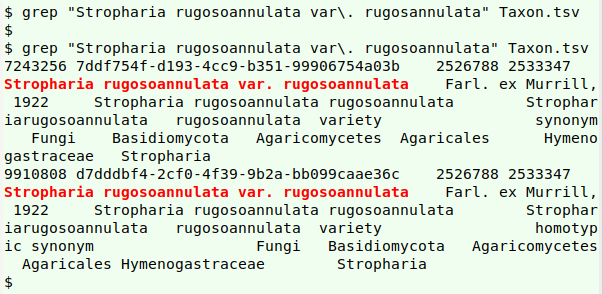
Advanced name-checking. Checking name strings composed of more than word (like "Stropharia rugosoannulata var. rugosoannulata Farl. ex Murrill, 1922") involves splitting the name into its component strings, discarding the unneeded bits ("var." and "Farl. ex Murrill, 1922"), then doing one-word checks on the remaining parts. Fortunately, there's already a fast and reliable command-line program that does the name-splitting and -discarding: gnparser, about which I've written before.
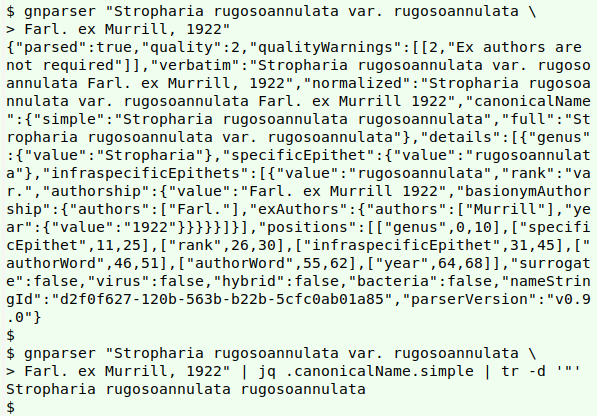
To get the simplest version of a complicated name string, just throw the name at gnparser and from its JSON output extract "canonicalName" and "simple" with jq, then delete the quotes from the JSON:
gnparser "Stropharia rugosoannulata var. rugosoannulata Farl. ex Murrill, 1922" | jq .canonicalName.simple | tr -d '"'
gnparser also works with lists of names:

So, to process a field with full scientific names, I do a one-name check on what gnparser | jq produces from the field (without its header), after converting spaces in the output to newlines. The new function (again for a tab-separated table) is:
2+namechk() { awk 'FNR==NR {a[$0]; next} $0 != "" && !($0 in a) {print $0}' ~/scripts/gbtnames <(tail -n +2 "$1" | cut -f"$2" | gnparser | jq .canonicalName.simple | tr -d '"' | tr ' ' '\n') | sort | uniq; }
In my real-world table "taxon.txt", the full, multi-word version of names is in field 3 ("scientificName"). The new function finds just one misspelling in field 3:

As shown in a screenshot above (the one with added blue underlines), the misspelling "projectullus" isn't in field 3, nor are the other errors I found earlier:
The functions "1namechk", "1namechkR" and "2+namechk" are now in my data auditing toolkit, and despite the size of the GBT names dictionary, the functions work very fast.
Last update: 2020-05-06
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License