
For a full list of BASHing data blog posts, see the index page. ![]()
Parsing scientific names
Personal names are data processing nightmares. Because the same personal name can be written in several different ways, name strings can be very hard to parse into their separate components. This hasn't stopped coders from trying, and you'll find lots of interesting products and projects by googling "name parser".
Scientific names for organisms are even harder to parse than personal names. The usual usage in zoology is a Latinised, 2-part animal name followed by the surname of the person who named the animal, followed by a comma and the year the name was first published by that person. Here's an example, broken into its parts:
Tasmanodesmus hardyi Chamberlin, 1920
Genus = Tasmanodesmus
species = hardyi
Author = Chamberlin
year = 1920
There are a number of complications. For example, the name might include a subgenus name in brackets or/and a subspecies name:
Cladethosoma (Haplethosoma) gladiator Jeekel, 1982
Genus = Cladethosoma
Subgenus = Haplethosoma
species = gladiator
Author = Jeekel
year = 1982
Heterocladosoma transversetaeniatum perarmatum Jeekel, 1987
Genus = Heterocladosoma
species = transversetaeniatum
subspecies = perarmatum
Author = Jeekel
year = 1987
Another wrinkle in zoology is that if the species is reclassified and put in a different genus, the surname of the original species name author is attached to the scientific name in brackets:
Hoplatessara luxuriosa (Silvestri, 1895)
Genus = Hoplatessara
species = luxuriosa
original Author of species name = Silvestri
year species name published = 1895
The name published by Silvestri in 1895 was "Strongylosoma luxuriosum"
Scientific names in botany are a bit different from zoological ones and have additional complications. An example from Australia:
Grevillea pyramidalis subsp. leucadendron (A.Cunn. ex R.Br.) Makinson
Genus = Grevillea
species = pyramidalis
subspecies = leucadendron
Author string = (A.Cunn. ex R.Br.) Makinson
"This name reflects the view of Alan Cunningham in a book published by Robert Brown in 1830, and its status in the opinion of Bob Makinson in 2000 published in the Flora of Australia"
As if scientific name parsing hadn't been made difficult enough by the naming rules, there are also disagreements about nomenclature
Should the Willow Tit be called Poecile montana or Poecile montanus?
plus variations in author name spelling, and typographic and other errors in the texts being parsed. Despite these challenges, a number of scientific name parsers have been developed over the years and all are more or less successful.
The latest parser I've seen is gnparser in GitLab, written in Go by Dmitry Mozzherin and Geoff Ower. The self-contained executable is available for Linux, has an MIT license and works on the command line. I looked at version 0.5.1.
Mozzherin warns that "the project is fresh out of the oven...and as such will go through a few changes in output before it settles". Users are enouraged to submit bug reports to the project's GitLab site if they see something is not working, or to dmozzherin@gmail.com if they don't have a GitHub or GitLab account.
gnparser can parse single names or long lists of names, and it's very fast. I wanted to try it out in my data auditing not so much for its ability to split up a scientific name into its components, but because gnparser also assesses name string "quality". gnparser gives each name it parses a quality score as follows:
"quality": 1 - No problems were detected
"quality": 2 - There were small problems, normalized result should still be good
"quality": 3 - There were serious problems with the name, and the final result is rather doubtful
"quality": 0 - A string could not be recognized as a scientific name and parsing fails
The output of gnparser is in JSON format, which isn't much fun for us human readers:
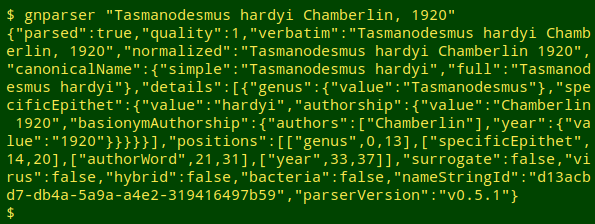
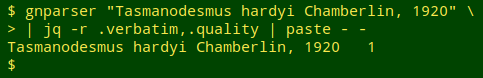
I use the command-line program jq to extract particular items of interest from JSON mumbo jumbo. For each name being audited I wanted the raw "verbatim" value and the raw "quality" value, which I could extract with jq's -r option. To put those 2 results on one line I piped the output to paste - -:
In a trial run I made up a list of incorrect variants and ran gnparser on the list:
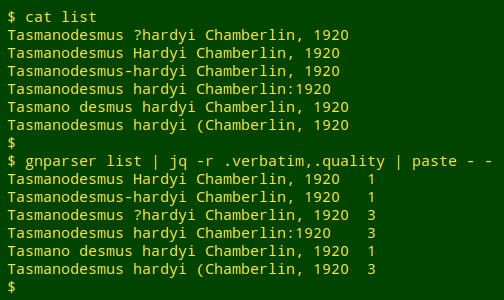
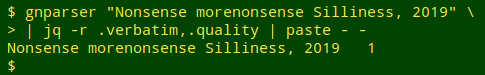
Notice that gnparser accepted 3 of the incorrect variants because they were well-formed. The full JSON output showed that the first "1" string was parsed as the genus "Tasmanodesmus" authored by "Hardyi Chamberlin", the second as the hyphenated single name "Tasmanodesmus-hardyi", and the third as the subspecies "hardyi" of species "desmus". In fact, gnparser will report "quality = 1" for just about any well-formed name string:
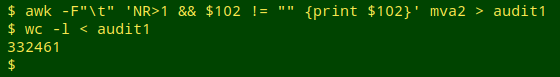
The tab-separated file to be audited, "mva2", had 355,824 records (plus a header) with scientific names in field 102. I first used AWK to pull out as "audit1" all the non-header lines in which field 102 wasn't blank. The result was a list of 332,461 scientific names (with many duplicates):
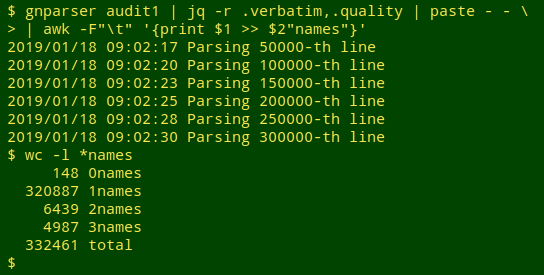
I then fed "audit1" to gnparser, jq, paste and an AWK command that would put the "quality = 1" names in a file called "1names", the "quality = 2" names in a file called "2names", and so on. gnparser helpfully reported where it was up to during the parsing process, which only took a few seconds:
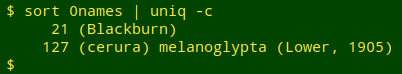
The 148 "quality = 0" name strings that caused parsing to fail were actually repeats of just 2 improperly formed ones:
Similarly, the 4,987 "quality = 3" strings were 111 unique name strings. Most had malformed authors or dates, but a few had name issues:
Examples:
Abantiades hyalinatus (Herrich-Schaffer, [1853])
Cardiochiles pollinator Dangerfield & A
Chelepteryx collesi 1835
Chimarra ct 268
Dichromodes sp
Hydrobiosella pt584
Pinara divisa group (Walker, 1855)
Unnamed 2
Just 88 unique strings made up the 6,439 "quality =2" set, about which the gnparser developers say "There were small problems, normalized result should still be good". As a zoological taxonomist I'd have to disagree about some of those, but overall the sorting was OK:
OK, interpretable:
Amphylaeus (Amphylaeus) Genus (Subgenus)
Anochetus armstrongi McAreavey J, 1949 Author name should be "J. McAreavey"
Not OK:
Acontiine (Angas) "Acontiine" not a name - refers to a species within Acontiinae (owlet moth subfamily)
Machomyrma (Tenison-Woods) Author name in brackets can't apply to genus name only
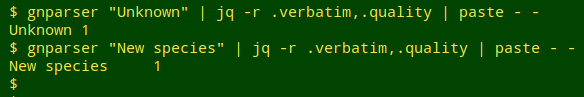
As a last check I went through the "1names" file and turned up what might be called false negatives, but there weren't many. They were strings like "Arctiid", which is not a name but means "species in the family Arctiidae". Fortunately there were no strings in "mva2" like "Unknown" or "New species":
I'm pretty happy with the latest gnparser and have added it to my data auditing toolkit.
Last update: 2019-01-20
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License