
For a full list of BASHing data blog posts, see the index page. ![]()
Comparing fields across two tables
It's a shell-user's axiom: if you find yourself typing certain commands again and again, script them.
The data-auditing job I was doing (again and again) was to compare the same field in two different tab-separated tables, looking for differences in the "same" data items. Below are two example tables. This is "fileA":
| localID | lastName | firstName | |
| RolKiC0h | Albrecht | Rosie | albrecht@yahoo.com |
| k1QZrQIu | Laursdatter | Zeynep | zeyneplaursdatter@gmail.com |
| TRcdUSJC | Torres | Alex | alextorres@gmail.com |
| 9dYfqWpZ | Fournier | Malo | malo.fournier@gmail.com |
| MDCZit6k | Rault | Alpinien | alpinienrault@laposte.net |
| ueJ7isSW | Yildirim | Aaron | aaron.yildirim@yahoo.com |
| rVoJHkp0 | Martinez | Enrique | martinez@telefonica.es |
| CjJuijEO | Osińska | Marta | osinska@poczta.onet.pl |
and this is "fileB":
| donation | localID | |
| 5 | sagalundquist@hotmail.se | DEn1MoBz |
| 10 | zeyneplaursdatter@gmail.com | k1QZrQIu |
| 15 | aaron.yildirim@yahoo.com | ueJ7isSW |
| 10 | emartinez2129@gmail.com | rVoJHkp0 |
| 20 | alpinienrault@laposte.net | MDCZit6k |
| 10 | malo.fournier@gmail.com | 9dYfqWpZ |
| 10 | saunier@free.fr | AjO6M0Tx |
| 15 | albrechtrosie4@gmail.com | RolKiC0h |
The fake names and emails are modified (in part) from the wonderful datafairy website. The localIDs were built with head -n3 /dev/urandom | tr -cd '[:alnum:]' | cut -c -8.
Each person in these tables has a unique localID. The question is, is the person's email address different in the two files? With these two tables, you can answer that question just by looking (there are two changes of email address), but the files I audit might have 100+ fields and several million records. Finding differences by eye is out of the question. To look for differences between large files I use a 3-step strategy, described below.
Pick and check a unique identifier (UI) field. In the example, the localID field will do this job. Unfortunately, in some of the data tables I audit, the "unique" ID isn't unique at all because of duplication or pseudo-duplication of records. After selecting a candidate UI field, I always check it for uniqueness. I first get a numbered list of fields from the header line of each table
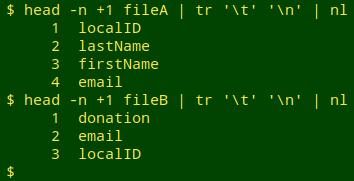
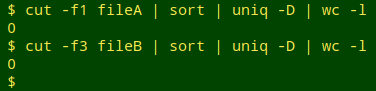
and then use the field numbers to cut out the UI fields (here, localID) from the two tables and check them for uniqueness. If the line count of duplicates is greater than zero in either table, I try to fix the duplication or pseudo-duplication problem before going any further. Note also that the name of the UI field is the same in the examples, but it might be different in the two tables I'm comparing in an audit.
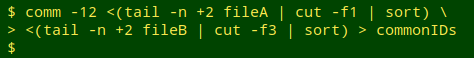
Select out the common records. Next, I build a list of the localIDs shared by the two tables, using comm -12. The list is saved as the temp file "commonIDs".
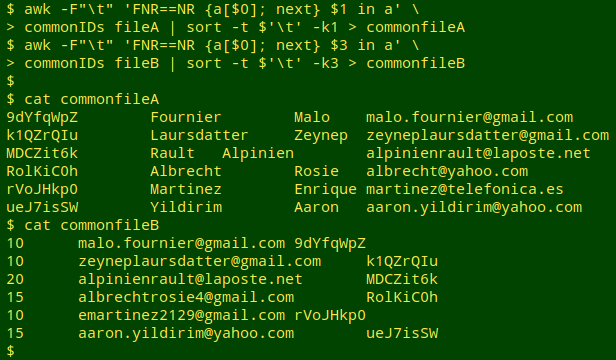
I then use the list of common IDs to select comparable records in the two tables. For small files this could be done with grep -f, but as I showed in an earlier BASHing data post, AWK is much quicker with big files. In the same command chain, the tables (now without header lines) are sorted by their localID fields.
Test for field differences. Now for the field comparison, which I do with AWK after paste-ing together one of the localID fields and the two fields to be compared. I first count the lines where there are differences. If there aren't many such lines, I print the results to screen.
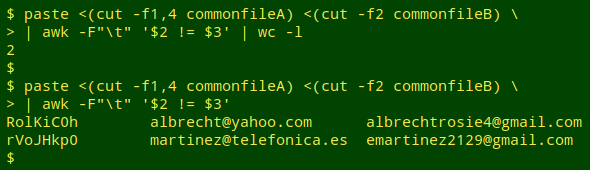
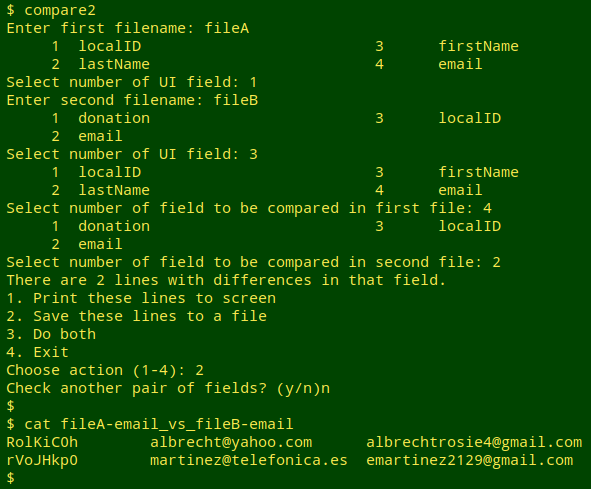
Scripting. That's the overall strategy, and below is the rough-as-guts script, "compare2", that's based on it. The working directory for the script is the one containing the two files to be compared. The script allows me to compare a series of field pairs in succession, either printing each to screen, saving each as an appropriately named file, or both.
#!/bin/bash
read -p "Enter first filename: " file1
flds1=$(head -n 1 "$file1" | tr '\t' '\n' | nl | pr -t -2)
echo "$flds1"
read -p "Select number of UI field: " ui1
uniqchk1=$(cut -f"$ui1" "$file1" | sort | uniq -D | wc -l)
if [ "$uniqchk1" -gt 0 ]; then
echo "That UI field contains duplicates. Exiting." && exit
else
read -p "Enter second filename: " file2
flds2=$(head -n 1 "$file2" | tr '\t' '\n' | nl | pr -t -2)
echo "$flds2"
read -p "Select number of UI field: " ui2
uniqchk2=$(cut -f"$ui2" "$file2" | sort | uniq -D | wc -l)
if [ "$uniqchk2" -gt 0 ]; then
echo "That UI field contains duplicates. Exiting." && exit
else
while true; do
comm -12 <(tail -n +2 "$file1" | cut -f"$ui1" | sort) \
<(tail -n +2 "$file2" | cut -f"$ui2" | sort) > /tmp/commonIDs
awk -F"\t" -v U="$ui1" 'FNR==NR {a[$0]; next} $U in a' \
/tmp/commonIDs "$file1" | sort -t $'\t' -k"$ui1" > /tmp/common"$file1"
awk -F"\t" -v U="$ui2" 'FNR==NR {a[$0]; next} $U in a' \
/tmp/commonIDs "$file2" | sort -t $'\t' -k"$ui2" > /tmp/common"$file2"
echo "$flds1"
read -p "Select number of field to be compared in first file: " fld1
echo "$flds2"
read -p "Select number of field to be compared in second file: " fld2
diffs=$(paste <(cut -f"$ui1","$fld1" /tmp/common"$file1") \
<(cut -f"$fld2" /tmp/common"$file2") | awk -F"\t" '$2 != $3')
diffscount=$(echo "$diffs" | wc -l)
echo "There are $diffscount lines with differences in that field.
1. Print these lines to screen
2. Save these lines to a file
3. Do both
4. Exit"
read -p "Choose action (1-4): " foo
case $foo in
1) echo "$diffs";;
2) echo "$diffs" > "$file1"-$(head -n 1 "$file1" | cut -f"$fld1")_vs_"$file2"-$(head -n 1 "$file2" | cut -f"$fld2");;
3) echo "$diffs" && echo "$diffs" > "$file1"-$(head -n 1 "$file1" | cut -f"$fld1")_vs_"$file2"-$(head -n 1 "$file2" | cut -f"$fld2");;
4) rm /tmp/common* && exit;;
esac
read -p "Check another pair of fields? (y/n)" zaz
case $zaz in
n) rm /tmp/common* && exit;;
y) continue;;
esac
done
fi
fi
rm /tmp/common*
exit
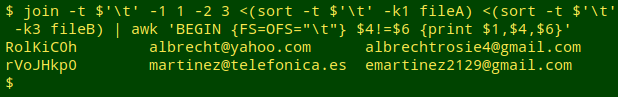
Update. Jean-Laurent Terrosi suggests an alternative approach using the join command. Here's an example using the same files as above:
join -t $'\t' -1 1 -2 3 <(sort -t $'\t' -k1 fileA) <(sort -t $'\t' -k3 fileB) | awk 'BEGIN {FS=OFS="\t"} $4!=$6 {print $1,$4,$6}'
Last update: 2019-02-08
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License