
For a full list of BASHing data blog posts, see the index page. ![]()
Bird watching with AWK and grep
What I call a "fileA-in-fileB" search is a search of fileB for lines that match any of the lines in fileA. There are a number of grep vs AWK speed tests on the Web for one or two fileA-in-fileB searches, but I thought I'd post what happens when you systematically vary a fileA-in-fileB search. TL;DR: AWK wins.
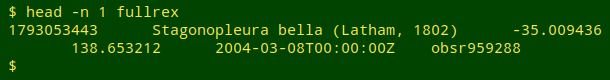
I'll demonstrate using bird sightings from Australia as recorded by the "eBird" program; the data come from the Global Biodiversity Information Facility. I trimmed the full, tab-separated data table of 10,132 626 sightings to the fields gbifID (a unique ID number for the sighting), scientificName (of the bird sighted), decimalLatitude, decimalLongitude, eventDate and recordedBy (code for the bird observer). I then shuffled the records. A typical record in the 1 GB "fullrex" file of sightings looks like this:
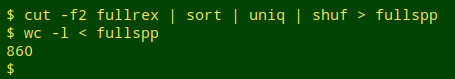
The scientificName field includes 860 different bird species and subspecies. I built a list of the 860 names and shuffled it as the file "fullspp":
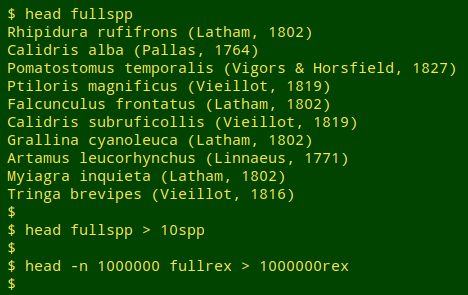
To show how I'll be testing the searches, I'll first make reduced samples of "fileA" (the bird species) and "fileB" (the sightings). The file "10spp" has just the first 10 names on the list of 860, and the file "1000000rex" has the first 1 million lines from the list of 10,132,626 sightings.
Three "fileA-in-fileB" searches and their elapsed times are shown below.
The searches were done on a desktop computer with an Intel (quad) i5-4590T processor running at 2 GHz, and 8GB of 1600 MHz DDR3 RAM. Note that the searches give identical results, but to prove that, it's necessary to do some sorting because the second method "interleaves" the lines it finds. Versions: GNU bash 4.4.12, GNU grep 2.27 (with no colouring), GNU Awk 4.1.4.
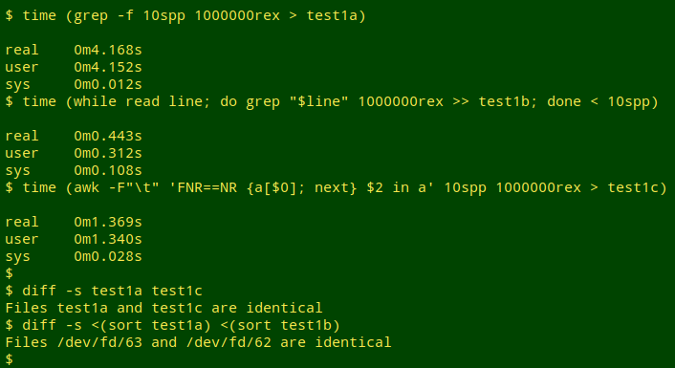
In the first search, grep -f looks for matches in "1000000rex" to the 10 lines in "10spp".
In the second search, the lines in "10spp" are fed one by one to grep in a while, read loop, with each successful match being appended to the output file.
In the third search, "10spp" is loaded into an AWK array. AWK then looks for lines in "1000000rex" in which the second field (bird species names) is a match for anything in the array.
The tests were repeated with the same 1 million records, but with 20, 50, 100, 200 and 500 species names. Here's a double-logarithmic (log-log) plot of the results:
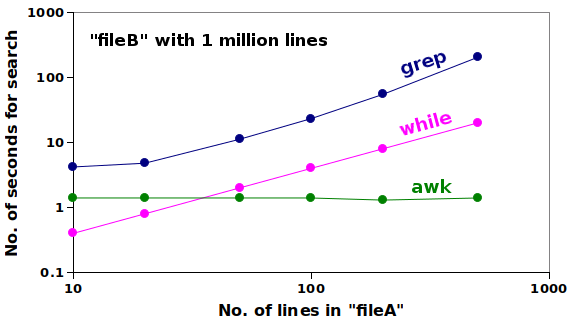
The grep -f method was always the slowest, and both grep -f and the while, read loop took increasingly longer to search as the size of "fileA" increased. AWK, on the other hand, took a constant 1.4 seconds for the search, even when the size of "fileA" increased 50-fold.
In the second test, I kept "fileA" at 100 lines, but searched 2 million, 5 million and 10 million records in "fileB", with the results shown in this log-log plot:
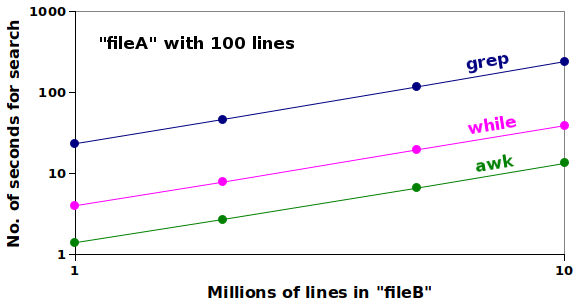
The search time increased for all three methods in proportion to the number of records searched, and the AWK array method was the fastest. Not only was AWK the speed champion, it was also a targeted searcher. It didn't look for the species name anywhere in the record, but only in the appropriate field.
Last update: 2018-10-24
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License