
For a full list of BASHing data blog posts, see the index page. ![]()
Fuzzy matching in practice
One of my tasks as a data auditor is to find approximate variants of certain data items, like the many versions of "sea level" I discovered in an elevations field, as described in an earlier BASHing data post.
My preferred command-line tool for this fuzzy-matching job is tre-agrep. It's available in the repositories and has a clearly written manpage. In this post I outline a practical method for using tre-agrep that suits my auditing work.
The demo file ("idby") is one complete field from a table of 36668 museum specimen records. Each line in the file is either blank or contains a string of one or more personal names, in one of several formats and combinations. The "idby" file is particularly messy and has spelling and other errors as well as format variants.
I first run the tally function from A Data Cleaner's Cookbook. Scanning the output gives me an overview of the names represented by approximate variants:
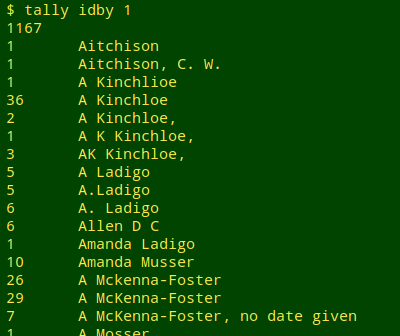
From the scan it's obvious that "Santangelo" (not shown in the screenshot above) has the largest number of variants, so I run the following command:
tre-agrep -is Santangelo -5 idby | sort | uniq -c | sed 's/$/\x1B[1;31m|\x1B[0m/'
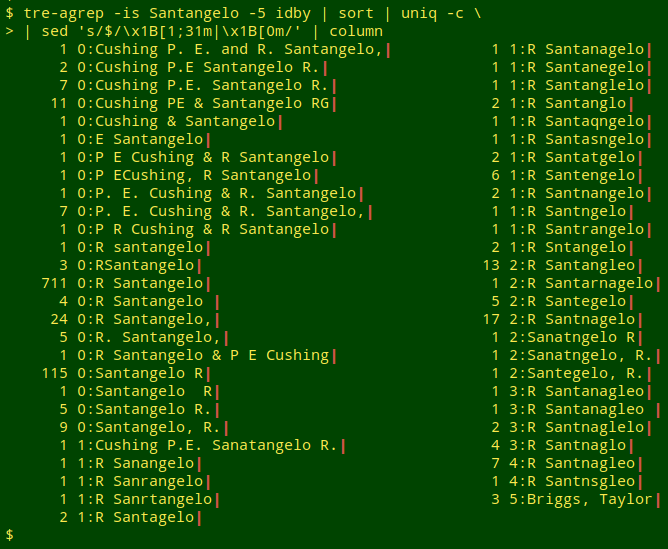
The "-i" option tells tre-agrep to ignore case, which is just as well, since one of the variants is "R santangelo". The "-s" option means that tre-agrep will report the "cost" involved in looking for approximate matches for "Santangelo". "Cost" includes character deletions, character insertions and character substitutions. The "-5" option is my choice for the maximum number of deletions, insertions and/or substitutions that tre-agrep will allow in its search. Why 5? I'll explain shortly.
The output from tre-agrep -s is the cost, a colon and the contents of the line found with the approximate match. For example, "1:R. Sanangelo" is an approximate match with a cost of 1, because 1 character has been deleted from "R. Santangelo".
The tre-agrep output is passed to sort, which sorts it in order of increasing cost, and then to uniq -c, which tallies up the unique results. Finally, I pass the result to sed, which prints the line ending ("$") as a red pipe to make it easier for me to see trailing spaces; see for example the 711 "R Santangelo" and the 4 "R Santangelo ".
The last unique approximate match, with a cost of 5, is "Briggs, Taylor" (see screenshot) and can obviously be ignored. I find in practice that 5 is a good default for maximum cost and captures all the variants in messy data. With a maximum cost of 5 and larger, the results can be very noisy with non-matches. In tidier datasets, I can drop the maximum cost to get a shorter output from the command chain.
This handy command chain can be saved as a function, which I'll call "fuzzy". It takes as arguments the file, the search term and the maximum cost:
fuzzy() { tre-agrep -is "$2" -"$3" "$1" | sort | uniq -c | sed 's/$/\x1B[1;31m|\x1B[0m/'; }
Having dealt with "Santangelo", I would next run tre-agrep on the second biggest set of variants in "idby", and so on.
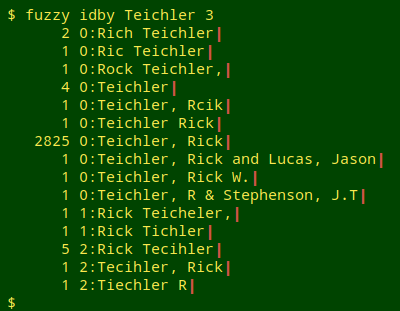
What happens next would depend on whether I was cleaning as well as auditing the data. The data source might advise, for example, that all 52 "Santangelo" versions could be normalised to "R. Santangelo" and "P.E. Cushing and R. Santangelo".
Data cleaning is a slow process, especially with files as messy as "idby", but I find that tre-agrep speeds up my work considerably.
The "clustering" feature in OpenRefine can be handy for approximate matching in some datasets. My experience with clustering in OpenRefine is that it's less flexible and harder to use than tre-agrep. And do I really need a large, complex application (80+ MB download) that runs in a browser and struggles with very large datasets, when I can do the same job on any size file with simple command-line tools?
Last update: 2020-01-13
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License