
For a list of BASHing data 2 blog posts see the index page. ![]()
Data entry with unknown data categories
Both Microsoft Excel and Google Sheets allow users to write data entry forms. The forms aren't very easy to create, but they help data workers to avoid entry errors when compiling data in spreadsheets.
Data entry forms require that the structure of the records is already known. What if you don't know in advance all the possible "columns" you might need?
That's the situation I'm in when I'm asked to identify the species in a set of biological samples. My final spreadsheet-able table will have samples as rows and species as columns, but I can't lay that out beforehand because I don't know which species will turn up. Each sample-species "cell" in the table, furthermore, will have a count of specimens or some biological notes.
My solution is to build a plain text file as I do my identifications, with semicolons and commas as temporary data-item separators. It's wonderfully jumbled: there's no special order to the samples I look at or the species I identify. Here's a simplified example, a file I'll call "jumble":
id002;sp2,6;sp1,4M+2F
id001;sp1,3J;sp5,12;sp2,7
id003;sp5,43
id006;sp3,4M;sp2,3;sp5,9
id004;sp1,2F;sp3,5J
id005;sp4,2;sp1,3M;sp5,6;sp3,1F
When all the identifying and data entry is done, I use the command line to bring order out of this not-really-chaos.
The first step is to use AWK to create comma-separated triplets with sample, species and count/note in that order:
awk -F";" '{for (i=2;i<=NF;i++) print $1","$i}' jumble
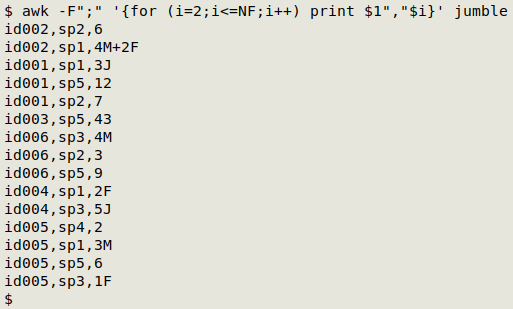
AWK divides each line into fields separated by a semicolon ( -F";"). It then processes each line field by field beginning with the second field (for (i=2;i<=NF;i++)). For each of these species-comma-count/note fields AWK first prints the sample field (print $1) followed by a comma, followed by the species-comma-count/note (print $1","$i).
This triplet stage is also a good one for checking to make sure there aren't any formatting errors in the data. If everything looks OK, I pass the result to GNU datamash to build a neatly sorted pivot table of samples vs species. I use sed to label the first, or samples "column":
datamash -t"," --filler="-" crosstab 1,2 collapse 3 | sed '1s/^/Sample/'
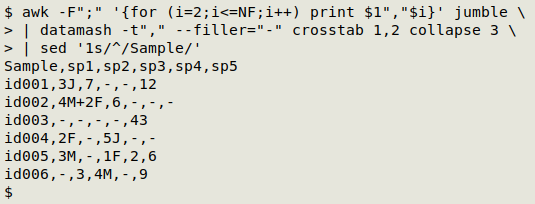
datamash is told that the field delimiter is a comma -t"," and that any missing values in the pivot table should be replaced with a hyphen (--filler="-"). The pivot table-building is specified with "crosstab 1,2", which means field 1 (sample) by field 2 (species). Entries in the pivot table should just be whatever is in field 3 (count/note): "collapse 3".
That finishes the job of building a spreadsheet-able CSV, but I can pass the result to column to give me a neat table in my terminal:
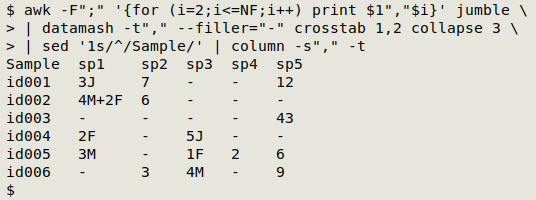
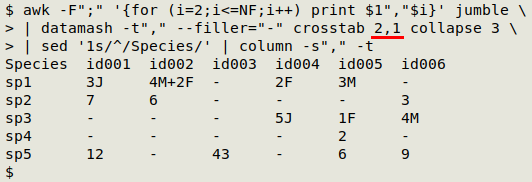
And swapping the datamash crosstab numbers and heading the first column with "Species" gives me a species-by-sample table from the same original jumble:
Next post:
2025-07-11 The script command for tinkerers
Last update: 2025-07-04
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License