
For a list of BASHing data 2 blog posts see the index page. ![]()
Munging the Atlas of Living Australia table format
I had a TSV (occurrence.tsv) with 182 fields but no header line and with every data item quoted. The 182 field names were in a separate file, meta.xml. To build a TSV (called "ocala") with a header line and without the quotes I wrote a function:
alaprep() { cat <(sed 's|taxonRankID|/taxonRankID|' meta.xml | awk -v FS="/|\"/>" '/field index="0"/ {f=1} f {printf("%s\t",$(NF-1))} /field index="181"/ {exit}' | sed 's/\t$/\n/') occurrence.tsv | sed 's/^"//g;s/"\t"/\t/g;s/"$//' > ocala; }
The overall structure is "concatenate (something-based-on-meta.xml) with occurrence.tsv, then delete all quotes around individual data items". The "something-based-on-meta.xml" is the missing header. Here are screenshots of parts of meta.xml:
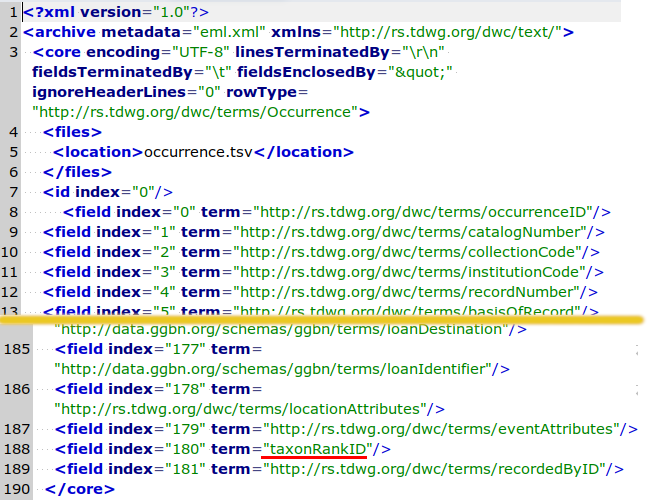
The linesTerminatedBy=="\r\n" bit is a fib; all lines had simple \n endings. Note that the fields are numbered from 0 to 181, and that the field names are at the ends of the URLs. For example, field 1 (index 0) in the TSV is occurrenceID. Note also that field 181 (index 180) has the only field name not preceded by a forward slash, taxonRankID.
The function works like this:
- Put a forward slash on that field 181 entry in meta.xml with sed:
sed 's|taxonRankID|/taxonRankID|' meta.xml - Pass the result to an AWK command that prints every field name followed by a tab (more on this command in a moment).
- Pass the AWK command output to sed to delete the trailing tab:
sed 's/\t$/\n/' - Concatenate the result of this chain with occurrence.tsv
- Pass occurrence.tsv, now with a header line with field names, to sed to delete line-beginning quotes, quotes around tabs and line-ending quotes:
sed 's/^"//g;s/"\t"/\t/g;s/"$//'
The AWK command treats both forward slashes and (escaped) quotes followed by forward slashes followed by right-pointing angles as field separators (-v FS="/|\"/>".
The command starts its work with the line beginning with field index="0" (/field index="0"/). Its first action is to set a flag "f" ({f=1}). The next action is to print the next-to-last field in the line followed by a tab (f {printf("%s\t",$(NF-1))}), if the condition "f" exists, and it does exist because of the first action. That next-to-last field is the one between field separators / and "/>; the last field is empty and follows the field separator "/>.
AWK continues this run until it reaches line 189 (/field index="181"/). Since the flag "f" is still set, it prints the field name and a tab, then does the requested action {exit} and closes.
And why did I have to add a header to a headerless TSV and delete zillions of unnecessary quotes? Well, the meta.xml and occurrence.tsv files came from the Atlas of Living Australia, and ALA is a bit strange with data (see below).
The background to this story begins with the Global Biodiversity Information Facility (GBIF). It's a clearinghouse for biological records, especially occurrences: a particular species ("what") was seen or collected at a particular spot ("where") on a particular day ("when") by a particular recorder ("who"). At the time I'm writing this, GBIF has so far more than 3 billion "what-where-when-who" occurrences.
GBIF gets records to index from sources ranging from museums and herbaria to citizen-science platforms like iNaturalist. It also gets records from organisations and projects that gather up data from lower-level recorders. One of these organisations is the Atlas of Living Australia (ALA).
To make data sharing easier, there's a globally accepted standard for "what-where-when-who" and other biological records. It's called Darwin Core and it has lots of standardised field names. For example, the "when" in a record can be slotted into verbatimEventDate, eventDate (the date or date/time in ISO 8601 format), year (number), month (number), day (number), and startDayOfYear and endDayOfYear (ordinal day numbers).
Some biological record contributors add their own, non-Darwin-Core fields, too, but these added fields should be explained in a metadata file, otherwise data users won't know what the field is supposed to be containing.
Contributors put their records on Web servers and tell GBIF where to find them. Occurrence records are nearly always shared with GBIF as simple TSVs in UTF-8 encoding, with Darwin Core field names in the header line. This is mainly because GBIF makes available free, open-source software for compiling records called the Integrated Publishing Toolkit (IPT). IPTs generate both the TSV and a metadata file, meta.xml, which lists the fields in the TSV's header line and references them to their source, such as the Darwin Core list of terms.
Are you still with me?
ALA doesn't use IPT software. It shares with GBIF tens of millions of records in its own, idiosyncratic way. The key components are an occurrence.tsv with no header at all and with every single data item surrounded with quotes, and a meta.xml file with numerous field-name reference URLs that don't resolve and one unreferenced field (taxonRankID) that no one uses, AFAIK, other than ALA.
For an example, go to this page on the GBIF website, click on the "Download" tab and choose "Source Archive". This sends you to an ALA server and returns a small ZIP archive with the aforementioned occurrence.tsv and meta.xml files.
Next post:
2024-12-13 Sorting camels, kebabs, pascals and snakes
Last update: 2024-12-06
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License