
For a full list of BASHing data blog posts see the index page. ![]()
DNA-style frameshift cryptography
Unless you've been asleep for a very long time, like a modern-day Rip van Winkle, you've probably learned that information in DNA is encoded in triplets of units that can be represented by the four letters A, C, G, and T. Each triplet of letters is a "codon".
What you may not have learned is that the codons in a DNA sequence can be read in any one of three ways, depending on where the triplet-reading starts:

From Wikipedia, showing forward reading's three possible reading frames. Original illustration by Hornung Ákos.
There can be more than one "correct" (biologically meaningful) reading of a sequence, too. DNA that can be triplet-read in more than one way is said to have multiple reading frames, and it's possible for two genes to share parts of the same DNA sequence. When these parts are triplet-read by a shift in where the reading starts, the two genes are said to have "out-of-phase overlaps". This Wikipedia article shows a nice example from the human genome.
Something similar could be done with multiple reading frames in order to encrypt a text. Instead of the four letters A, C, G and T, use just "0" and "1". Instead of a triplet as the unit of information, use an octet like "00110011" or "10000111", better known to us nerds as a byte. There are 64 different codons with A, C, G and T, but 256 different bytes. DNA can have three different correct readings in what's called the "sense" direction, but a string of 0s and 1s can be broken into contiguous bytes in eight different ways in either direction (left to right, and right to left), for a total of 16 different readings.
Further, some of the 256 bytes might encode different characters, even with non-scrambled encodings. For example, as mentioned in the last BASHing data post the character á is 11100001 in Windows-1252 encoding, but 10000111 in Mac OS Roman. Throw in some scrambled encodings and you have a simple but interesting cipher.
Let's see how that might work using the UTF-8 character set. To make the job easier, I'll use the space-separated table "utf8vals", shown below in six columns to save space.
To restore the original "utf8vals", copy and paste the table below into a text file, "pasted", then run tr '\t' '\n' < pasted | sort > utf8dvals.
| 00100001 ! | 00100010 " | 00100011 # | 00100100 $ | 00100101 % | 00100110 & |
| 00100111 ' | 00101000 ( | 00101001 ) | 00101010 * | 00101011 + | 00101100 , |
| 00101101 - | 00101110 . | 00101111 / | 00110000 0 | 00110001 1 | 00110010 2 |
| 00110011 3 | 00110100 4 | 00110101 5 | 00110110 6 | 00110111 7 | 00111000 8 |
| 00111001 9 | 00111010 : | 00111011 ; | 00111100 < | 00111101 = | 00111110 > |
| 00111111 ? | 01000000 @ | 01000001 A | 01000010 B | 01000011 C | 01000100 D |
| 01000101 E | 01000110 F | 01000111 G | 01001000 H | 01001001 I | 01001010 J |
| 01001011 K | 01001100 L | 01001101 M | 01001110 N | 01001111 O | 01010000 P |
| 01010001 Q | 01010010 R | 01010011 S | 01010100 T | 01010101 U | 01010110 V |
| 01010111 W | 01011000 X | 01011001 Y | 01011010 Z | 01011011 [ | 01011100 \ |
| 01011101 ] | 01011110 ^ | 01011111 _ | 01100000 ` | 01100001 a | 01100010 b |
| 01100011 c | 01100100 d | 01100101 e | 01100110 f | 01100111 g | 01101000 h |
| 01101001 i | 01101010 j | 01101011 k | 01101100 l | 01101101 m | 01101110 n |
| 01101111 o | 01110000 p | 01110001 q | 01110010 r | 01110011 s | 01110100 t |
| 01110101 u | 01110110 v | 01110111 w | 01111000 x | 01111001 y | 01111010 z |
| 01111011 { | 01111100 | | 01111101 } | 01111110 ~ | 10100001 ¡ | 10100010 ¢ |
| 10100011 £ | 10100100 ¤ | 10100101 ¥ | 10100110 ¦ | 10100111 § | 10101000 ¨ |
| 10101001 © | 10101010 ª | 10101011 « | 10101100 ¬ | 10101110 ® | 10101111 ¯ |
| 10110000 ° | 10110001 ± | 10110010 ² | 10110011 ³ | 10110100 ´ | 10110101 µ |
| 10110110 ¶ | 10110111 · | 10111000 ¸ | 10111001 ¹ | 10111010 º | 10111011 » |
| 10111100 ¼ | 10111101 ½ | 10111110 ¾ | 10111111 ¿ | 11000000 À | 11000001 Á |
| 11000010 Â | 11000011 Ã | 11000100 Ä | 11000101 Å | 11000110 Æ | 11000111 Ç |
| 11001000 È | 11001001 É | 11001010 Ê | 11001011 Ë | 11001100 Ì | 11001101 Í |
| 11001110 Î | 11001111 Ï | 11010000 Ð | 11010001 Ñ | 11010010 Ò | 11010011 Ó |
| 11010100 Ô | 11010101 Õ | 11010110 Ö | 11010111 × | 11011000 Ø | 11011001 Ù |
| 11011010 Ú | 11011011 Û | 11011100 Ü | 11011101 Ý | 11011110 Þ | 11011111 ß |
| 11100000 à | 11100001 á | 11100010 â | 11100011 ã | 11100100 ä | 11100101 å |
| 11100110 æ | 11100111 ç | 11101000 è | 11101001 é | 11101010 ê | 11101011 ë |
| 11101100 ì | 11101101 í | 11101110 î | 11101111 ï | 11110000 ð | 11110001 ñ |
| 11110010 ò | 11110011 ó | 11110100 ô | 11110101 õ | 11110110 ö | 11110111 ÷ |
| 11111000 ø | 11111001 ù | 11111010 ú | 11111011 û | 11111100 ü | 11111101 ý |
| 11111110 þ | 11111111 ÿ |
"utf8vals" holds the 188 visible characters and their binary values, out of the 256 single-byte encodings in UTF-8. I'll be using this table in an AWK array to convert bytes to characters.
The first demonstration string is the left-to-right "0001010101010001100011000101", which is the byte for capital "Q" preceded and followed by an invisible control character, and finished off with "0101". To retrieve the "Q" and replace non-visible characters with a hyphen, I can use the following command:
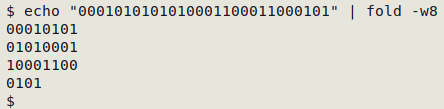
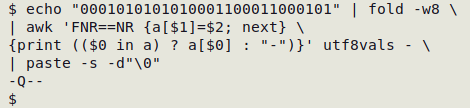
echo "0001010101010001100011000101" | fold -w8 \
| awk 'FNR==NR {a[$1]=$2; next} \
{print (($0 in a) ? a[$0] : "-")}' utf8vals - \
| paste -s -d"\0"
The fold -w8 command breaks the string into 8-character pieces, starting with the leftmost character, and puts them on separate lines:
The AWK command first builds an array from "utf8vals", with the binary string (field 1) as the index string and the corresponding character (field 2) as the value string. Note that no field separator needs to be specified, since a space is a field separator by default in AWK. With the array built, AWK moves to the broken-up demonstration string, here represented as the second argument "-", which means "standard input", and processes the 8-character pieces line by line. If the piece is an index string in the array, AWK prints the corresponding character. If not, AWK prints a hyphen.
The AWK output is passed to paste -s -d"\0", which puts the recovered characters (and hyphens) on a single line with no separator. Here's the result:
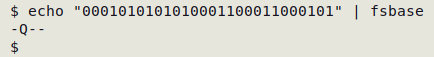
To save typing I can put the command in a function, "fsbase":
fsbase() { fold -w8 | awk 'FNR==NR {a[$1]=$2; next} {print (($0 in a) ? a[$0] : "-")}' utf8vals - | paste -s -d"\0"; }
Now for a file ("cryptic") with a secret message hidden inside it:
1110111001111110011010001001000010000100110001101100100111011110110111011100000010011110110001101100011011010100110000100100100010010111000001
To find the message I'll examine all of the 16 possible byte readings: 8 frames each in the forward and reverse directions. The command I'll use is a BASH for loop that has cut start the string (to be sent to "fsbase") at positions 1, 2, 3 etc. I'll also print a caption for each of the 16 results:
for i in {1..8}; \
do printf "forward, starting at position $i: "; \
cut -c "$i"- cryptic | fsbase; done
for i in {1..8}; \
do printf "reverse, starting at position $i: "; \
cut -c "$i"- <(rev cryptic) | fsbase; done
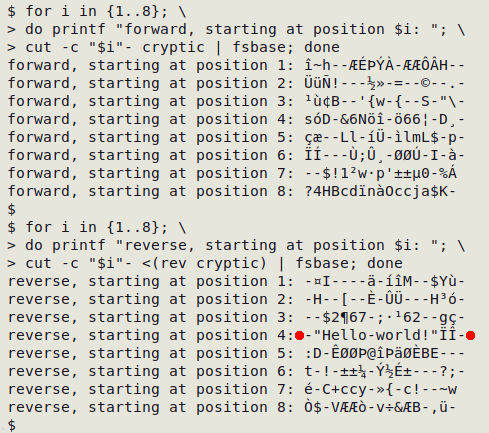
There's the secret message, revealed when converting bytes to characters in the right-to-left string after starting the 8-character "fsbase" operation at character 4.
OK, I found the message by eyeballing each of the 16 possible results. How do molecular biologists select the correct reading of a DNA sequence, out of the three possibles?
There isn't a simple answer. A DNA sequence that encodes a protein will have a "start" codon (ATG) at the beginning of the protein's code and a "stop" codon (TGA, TAG or TAA) at the end. Of the possible readings, the longest sequence between "start" and "stop" is regarded as the one most likely to be correct.
Last update: 2022-02-16
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License