
For a full list of BASHing data blog posts see the index page. ![]()
Hunting Excel date twins
Certain versions of Microsoft Excel for Mac counted days from 1 January 1904, while other Excel versions numbered their days from 1 January 1900. Microsoft calls these "the 1904 date system" and "the 1900 date system", and says there are problems you may encounter when you use workbooks that use different date systems.
I wrote about one of those problems in a 2017 post for The Linux Rain blog. If you compile a spreadsheet using a mix of "1900" and "1904" dates, you might have the same record represented twice, with dates exactly 4 years and 1 day apart (1462 days). These pseudo-duplicates are "Excel date twins".
The first twins I found (by accident) were in the Atlas of Living Australia (ALA) and had been imported from the Tasmanian Natural Values Atlas (TNVA). From the ALA they went to the Global Biodiversity Information Facility (GBIF). Both records say that I observed the peripatus species Ooperipatellus cryptus at a certain spot in Tasmania. One record says I did it on 3 February 1976, the other on 2 February 1972. The second date is an Excel date twin and impossible: I first came to Australia in January 1973.
Here are my twins:
ALA:
1976-02-03
https://biocache.ala.org.au/occurrences/94946259-785c-4f1a-ac14-a496ad4d719d
1972-02-02
https://biocache.ala.org.au/occurrences/1e633a0e-82a6-46d9-a7c4-46a99e2394cd
GBIF:
1976-02-03
https://www.gbif.org/occurrence/1647193398
1972-02-02
https://www.gbif.org/occurrence/1648019104
Guessing that the TNVA had more Excel date twins than mine, I had a go at looking for more of them in 2017. The code I used then was fairly clunky so I recently tried again with a slightly different approach, described below.
Preparing the data. I downloaded the TNVA dataset in GBIF as GBIF received it from ALA (the "source archive"). From the 1,121,933 records I pruned out those with no ISO 8601 date, with no latitude and longitude or with no observer (or observer unknown), saving the 807,895 remaining records in the header-less TSV "proc1".
There was a problem with "proc1". The same species might have been seen at the same spot on the same day by the same observer, but recorded twice. One observation might have been in the morning and another in the afternoon, or one observation was of a male and the other of a female. The result was several thousand species/location/observer/date duplicates in the TNVA. For the purpose of hunting Excel date twins these duplicates were irrelevant, so I trimmed "proc1" to the 5 fields
- scientific name of the species observed
- latitude in decimal degrees
- longitude in decimal degrees
- observer name string
- observation date in ISO 8601 format (YYYY-MM-DD)
and sorted and uniquified the result, generating the 794,323-record file "proc2".
What I wanted next was a 6th field with a serial day number corresponding to the observation date. To build this field my reference date was 1700-01-01 (a "1700 date system"). I chose that date because observation dates in TNVA go back to the 1700s, and I wanted all serial day numbers to be positive. The command to create the 6-field TSV "proc3" was:
awk 'BEGIN {FS=OFS="\t"} {split($5,a,"-"); \
$6=((mktime(a[1]" "a[2]" "a[3]" 0 0 0") \
- mktime("1700 01 01 0 0 0"))/86400); \
print}' proc2 > proc3
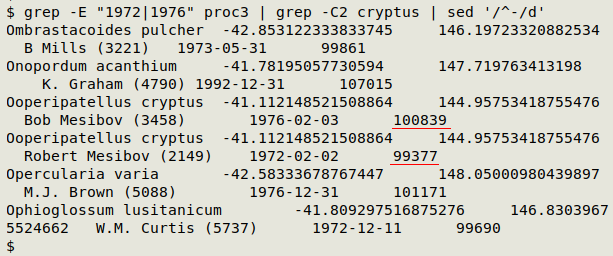
Here's a sample of "proc3" with my twin peripatus records. Note the difference in serial day number for my twins: 100839 - 99377 = 1462 days.
Getting some candidates. Next, I tweaked the 2017 strategy a little. First I added 1462 days to every serial day number in "proc3", building "proc3_1462":
awk 'BEGIN {FS=OFS="\t"} {$6=$6+1462} 1' proc3 > proc3_1462
I then cut out the scientific name, latitude, longitude and serial day number fields from "proc3" and "proc3_1462", sorted and uniqified them and passed them to comm -12 to print to "list" all the lines with the same entries in those 4 fields in both files.
comm -12 <(cut -f1-3,6 proc3 | sort | uniq) \
<(cut -f1-3,6 proc3_1462 | sort | uniq) > list
The resulting file "list" contained all the unique, abbreviated "proc3_1462" records that corresponded to unique, abbreviated "proc3" records with an added 1462 days. To get abbreviated "proc3" records without the added days, I rebuilt "list" with AWK, subtracting 1462 from the 4th field. I paired the candidate twins in the file "abbreviated_candidates":
awk 'BEGIN {FS=OFS="\t"} {print $0; $4=$4-1462; \
print $0"\n---"}' list > abbreviated_candidates
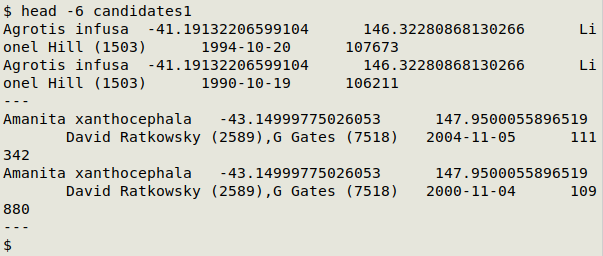
Here's the top of "abbreviated_candidates" in a text editor:

Getting observers into the candidates. Not all the paired records in "abbreviated_candidates" are genuine Excel twins: the 2 records might have had different observers. I excluded observers when building "list" because the TNVA sometimes uses different strings for the same observer. See the first screenshot above for an example, where I appear as "Bob Mesibov (3458)" in 1976 and as "Robert Mesibov (2149)" in 1972.
I added observers (and ISO 8601 dates) to "abbreviated_candidates" from "proc3" with an AWK command, to build "candidates1":
awk -F"\t" 'FNR==NR {a[$1 FS $2 FS $3 FS $6]=$4 FS $5; next} \
($0 in a) {print $1 FS $2 FS $3 FS a[$0] FS $4} /---/ {print $0}' \
proc3 abbreviated_candidates > candidates1
Checking for different observers. All the paired records in "candidates1" with the same observer string are likely to be genuine Excel date twins, and some of the others might also be. To find the record pairs with different observers I first put each triplet of lines (newer record, older record, "---" spacer) on a single line with paste, using the tab character as separator. I then used AWK to find lines where fields 4 and 10 (the 2 observer-string fields) were different, and "re-assembled" the resulting triplets with sed:
paste - - - -d "\t" < candidates1 \
| awk -F"\t" '$4 != $10' \
| sed 's/\t/\n/6;s/\t/\n/11' > possible_discards
A sample of "possible_discards" is shown below. The first 2 pairs are Excel date twins. The second 2 aren't because they're records from different observers.
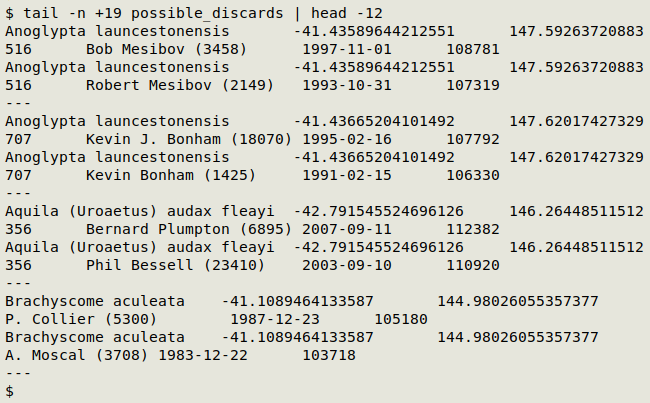
Short of building a lookup table with string equivalents like "Robert Mesibov | Bob Mesibov", the quickest way to find genuine discards among the possible ones was to do it by eye in a text editor. I scanned the 184 triplets and discarded those where I knew the observer was really the same person, building "genuine_discards" with 143 triplets.
Filtering the candidates. The last step was to remove from "candidates1" the "genuine_discards". I did this by again using the "single line" trick and AWK, and re-assembling with sed:
awk 'FNR==NR {a[$0]; next} !($0 in a)' \
<(paste - - - -d "\t" < genuine_discards) \
<(paste - - - -d "\t" < candidates1) \
| sed 's/\t/\n/6;s/\t/\n/11' > candidates2
The output file "candidates2" has 762 pairs of likely Excel date twins, which is a relatively small but impressive stuff-up courtesy Microsoft Corporation and the TNVA. I may have missed a few by screening out within-day and different-specimen duplicates at the beginning of the process. On the other hand, it's possible that in some cases the same species was really, truly observed by the same person at the same spot on two dates exactly 4 years and 1 day apart.
In any case, I wouldn't like the job of hunting Excel date twins within GBIF's current total (2021-02-27) of 1,658,379,221 occurrence records. The coding would need to be a little bit more sophisticated...
The quality of TNVA data is pretty dreadful, and the TNVA has another problem. There are 19 fields in the TNVA dataset in ALA that are populated (have data entries) but don't appear in GBIF.
I queried both GBIF and ALA about this difference in January 2020. GBIF replied that the 19 fields were missing from the data they received from ALA. ALA hasn't responded yet (2021-02-27).
Last update: 2021-03-09
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License