
For a full list of BASHing data blog posts see the index page. ![]()
Building a data table from a sentence
In an excellent 2018 article aimed at mammal-studying scientists, three specialists described how to make data more easily re-usable.
One of their not-easily-reusable examples is a list of specimens and their museum catalog numbers:
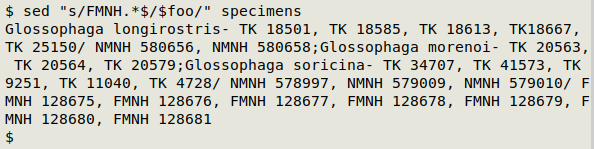
Glossophaga longirostris- TK 18501, TK 18585, TK 18613, TK18667, TK 25150/ NMNH 580656, NMNH 580658;Glossophaga morenoi- TK 20563, TK 20564, TK 20579;Glossophaga soricina- TK 34707, TK 41573, TK 9251, TK 11040, TK 4728/ NMNH 578997, NMNH 579009, NMNH 579010/ FMNH 128675 to FMNH 128681
In this single line of text (very slightly modified from the published example)
- species names are separated from catalog numbers with a hyphen and space
- catalog numbers are separated with a comma and space
- specimen collections are separated with a forward slash and space
- species data are separated with a semicolon
- a catalog number range is indicated with " to "
The line of text uses punctuation as a kind of data compression tool. Compression like this is typical of some scientific journals, and is meant to save space and ink. The data could be expanded into a delimited text file like this (I've used a pipe as field separator):
Glossophaga longirostris|TK 18501
Glossophaga longirostris|TK 18585
Glossophaga longirostris|TK 18613
Glossophaga longirostris|TK 18667
Glossophaga longirostris|TK 25150
Glossophaga longirostris|NMNH 580656
Glossophaga longirostris|NMNH 580658
Glossophaga morenoi|TK 20563
Glossophaga morenoi|TK 20564
Glossophaga morenoi|TK 20579
Glossophaga soricina|TK 34707
Glossophaga soricina|TK 41573
Glossophaga soricina|TK 9251
Glossophaga soricina|TK 11040
Glossophaga soricina|TK 4728
Glossophaga soricina|NMNH 578997
Glossophaga soricina|NMNH 579009
Glossophaga soricina|NMNH 579010
Glossophaga soricina|FMNH 128675
Glossophaga soricina|FMNH 128676
Glossophaga soricina|FMNH 128677
Glossophaga soricina|FMNH 128678
Glossophaga soricina|FMNH 128679
Glossophaga soricina|FMNH 128680
Glossophaga soricina|FMNH 128681
In this format the list of specimens examined could be made available as a separate, downloadable file by the scientific journal. But traditions die hard, alas, and in 2020 many journals still insist on barely usable data formats like that single line of text and don't offer the same data in a downloadable table. Fortunately, command-line enthusiasts like us can come to the rescue by reformatting. First I'll save that line of text as the file "specimens". Now, let's see...
I'll start by expanding the
FMNH 128675 to FMNH 128681
as
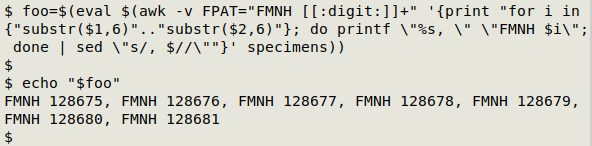
FMNH 128675, FMNH 128676, FMNH 128677, FMNH 128678, FMNH 128679, FMNH 128680, FMNH 128681
I can pull out the two catalog numbers by telling AWK that the field pattern in "specimens" is "FMNH " followed by some digits, then printing the two fields (GNU AWK 4 or later is needed for "FPAT"):
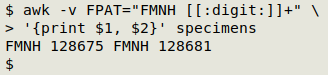
I can extract the numbers as substrings of those 2 fields:
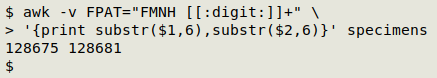
And I can get AWK to print out a BASH command containing those numbers in a range expression:
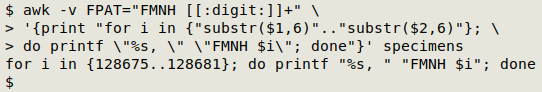
If I try running that printed-out command in the shell, I get just the expanded string of catalog numbers I wanted. However, the result's not quite perfect, because the output ends in ", " (underlined below in red). I can delete that ending with sed:
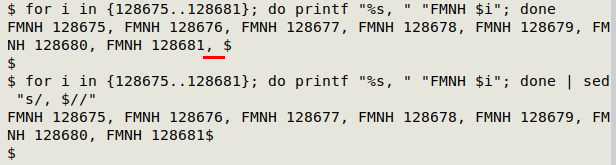
OK, now to run the full command in the shell using eval, and store the result in the shell variable "foo":
Next I'll use the expanded string as a replacement for "FMNH 128675 to FMNH 128681" in "specimens":
Finally, I'll reformat the single, expanded line of text as a pipe-delimited table. This is easy to do with AWK:
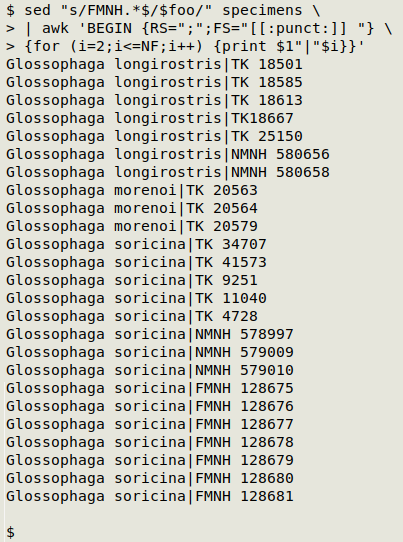
What's happening here is that AWK divides the string into 3 records, one for each species, because I've specified ";" as the record separator RS. Within each record, the field separator is any sort of punctuation (in this case it will be a hyphen, a comma or a forward slash) followed by a space (FS="[[:punct:]] "). This makes separate fields of the species name (field 1) and the various catalog numbers in each record. For each of the fields after the first one (for (i=2;i<=NF;i++)) I print the first field (the species name), a pipe and the selected field (print $1"|"$i).
A tidy-up tweak would be to remove that last, blank line, for which I could use "AWK NF" (print non-blank lines) or grep "." (search for lines containing any character). Here's the full reformatting code for "specimens":
foo=$(eval $(awk -v FPAT="FMNH [[:digit:]]+" '{print "for i in {"substr($1,6)".."substr($2,6)"}; do printf \"%s, \" \"FMNH $i\"; done | sed \"s/, $//\""}' specimens))
sed "s/FMNH.*$/$foo/" specimens | awk 'BEGIN {RS=";";FS="[[:punct:]] "} {for (i=2;i<=NF;i++) {print $1"|"$i}}' | awk NF
My hack requires two passes over "specimens". It could probably be done in one pass, but the code would look much uglier.
Last update: 2020-10-07
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License