
For a full list of BASHing data blog posts see the index page. ![]()
Finding malformed markup
When auditing data tables I sometimes stumble over a difficult problem with embedded HTML markup. The problem is nicely illustrated by the following real-world example. It's a single record from the tab-separated text file, "Refs":

The tags <i> and </i> were originally used to put an enclosed string in italics, maybe on a web page or in a program that understood HTML markup. The markup is correct around Brotia but is malformed around Zoosystematics and Evolution.
In "Refs" and similar files I've found other malformed tags, such as
</>
</em/>
<i/>
</i.</em>
<io>
<iW>
<i<[text]
<i[text]
<[text]</i>
<[text]</em>
<[spaces]i>
In addition to malformed tags, I've found correctly formed tags in the wrong place, such as in </i>some text<i>. I've also seen non-HTML markup in data tables, such as <italic>, <bold> and <roman>.
How to find malformed and misplaced markup efficiently on the command line? Checking every instance of < and > would be painfully slow, and I haven't yet found any validator or parser for HTML that can identify all these markup problems, mainly because I'm not actually auditing HTML documents.
My solution-in-progress isn't very efficient. The method has two stages and uses two small, reference files — "openers" and "closers" — listing the HTML tags I'm most likely to find in data tables. The files are newline-separated lists but are shown below as comma-separated files to save space:
The actual files are newline-separated lists!
"openers"
<b>,<B>,<em>,<EM>,<h1>,<H1>,<h2>,<H2>,<h3>,<H3>,<h4>,<H4>,<h5>,<H5>,<h6>,<H6>,<i>,<I>,<p>,<P>,<strong>,<STRONG>,<sub>,<SUB>,<sup>,<SUP>
"closers"
</b>,</B>,<br>,<br />,<BR>,<BR />,</em>,</EM>,</h1>,</H1>,</h2>,</H2>,</h3>,</H3>,</h4>,</H4>,</h5>,</H5>,</h6>,</H6>,</i>,</I>,</p>,</P>,</strong>,</STRONG>,</sub>,</SUB>,</sup>,</SUP>
(1) Check for correctly formed but misplaced tags. This first stage uses a while loop to read one of the reference files, then searches with grep for occurrences of tags that might be "improperly" adjoining alphanumeric characters or isolated by spaces on either side:
while read line; do grep -E "[[:alnum:]]$line|[[:blank:]]$line[[:blank:]]" file-to-be-checked; done < openers
while read line; do grep -E "$line[[:alnum:]]|[[:blank:]]$line[[:blank:]]" file-to-be-checked; done < closers
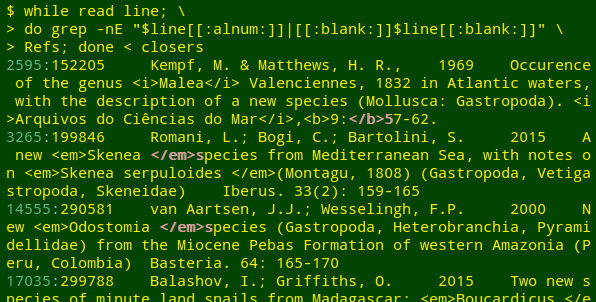
As you can see in the above screenshot, the first stage also finds awkward but valid placements of tags. In line 14555, for example, <em>Odostomia[space]</em> might be more neatly coded as <em>Odostomia</em>[space], but the two markups look the same to an HTML-interpreting program.
(2) Replace correctly formed tags and look for stray < and > in the modified file. The command is:
awk 'FNR==NR {a[$0]=$0; next} \
{for (i in a) gsub(a[i],"{MARKUP}"); print FNR": "$0}' \
<(cat openers closers) file-to-be-checked | grep -E "<|>"
An AWK command builds an array "a" from the concatenated "openers" and "closers" files. Moving to the file to be checked, AWK replaces all of the correctly formed tags in each line (if any) with the string "{MARKUP}". The line is then printed and preceded by its line number. The output from the AWK command is passed to grep for selection of any lines with < or > and highlighting of those two characters.
Here I've shown the first three positive results from "Refs" with grep --color=always to keep the grep highlighting, and I've underlined the finds:

My method is pretty kludge-y, but it detects all the problems I've so far noticed in the datasets I audit. If I find other types of markup errors I'll revise or supplement the code!
Last update: 2019-07-19
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License