
For a full list of BASHing data blog posts, see the index page. ![]()
Compare parts of strings
Here's the problem in a general form: in the following tab-separated list of strings, is there a quick way to find the lines where just the first word in the left-hand string differs from the first word in the right-hand string? Where just the second word differs? Where both words differ?

"Yes", three times: use AWK and its built-in split function.
The first command looks like this:
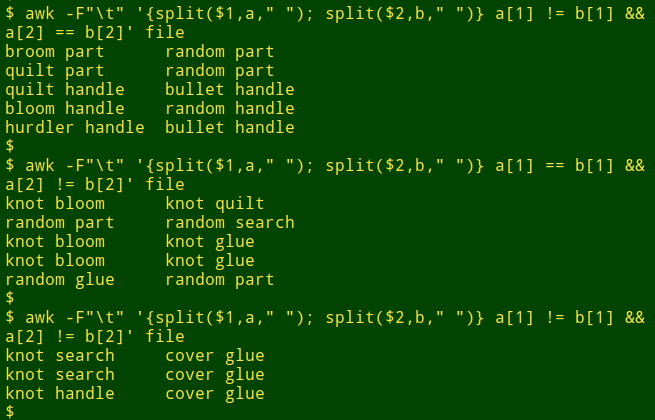
awk -F"\t" '{split($1,a," "); split($2,b," ")} a[1] != b[1] && a[2] == b[2]' file
The command first tells AWK that the field separator is a tab character (-F"\t"). In the action part of the command (between curly braces), split breaks up the first field ($1) into parts separated by a space (" ") and puts the parts in an array (a). The same thing happens to the second field ($2), but this time the parts go into a different array (b). Next, AWK looks for a pattern: lines where the first element in array "a" is different from (!=) the first element in array "b", and (&&) the second element in "a" is the same as (==) the second element in "b". The default action of AWK is to print any such lines it finds, so the command is complete.
And here are the results for only first word different, only second word different, and both words different:
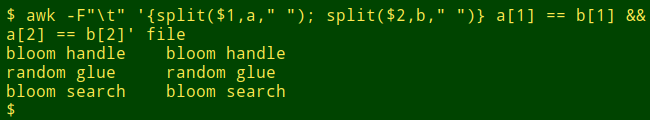
And lines where the left- and right-hand words are the same? Just edit the command a little:
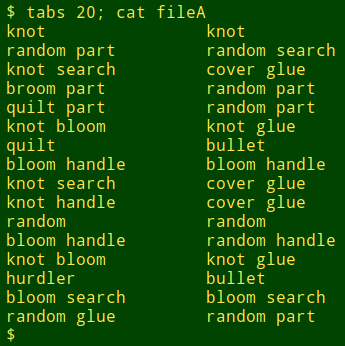
I used AWK and split in just this way recently when comparing lists of scientific names. In that case the first word was a genus name, the second word (if present) was a species name and the third word (if present) was a subspecies name. The "if present" condition was a complication, but AWK again offers a simple solution. Suppose I want to compare lines which only have genus names, to see where those differ. To mimic this I'll edit the words file a bit:
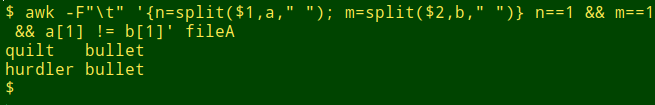
The command to check for "one-word-only" lines is shown below. In the action part I count the parts that split finds and store that count in the variable "n" for the first field and "m" for the second. I then add the additional conditions to the pattern that both "n" and "m" have to equal 1. In other words, AWK looks for lines where split only finds 1 part in each field.
awk -F"\t" '{n=split($1,a," "); m=split($2,b," ")} n==1 && m==1 && a[1] != b[1]' fileA
To look for two-word lines where the first words are different but the second words are the same, just filter lines where "n" and "m" are 2:
awk -F"\t" '{n=split($1,a," "); m=split($2,b," ")} n==2 && m==2 && a[1] != b[1] && a[2] == b[2]' fileA
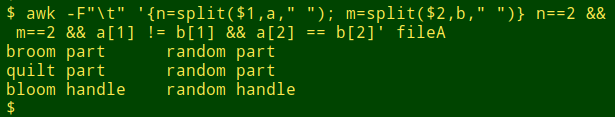
AWK compared several hundred thousand scientific names in a couple of seconds. To answer different questions all I had to do was "Up arrow" to the last-used command and edit it slightly. There are other command-line ways to do the "compare parts of strings" job, but this AWK method is particularly fast and simple.
Last update: 2018-05-22
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License