Between fields
one-to-many pairs
one2many triplets
Pairs and triplets
Between name fields
Between date fields
Between-field miscellany
one-to-many pairs
The one2many function looks for multiple values associated with the same unique value. As arguments it takes the filename, the number of the unique field and the number of the field being tested for multiple values.
one2many() {
awk -F"\t" -v one="$2" -v many="$3" 'NR==1 {print "No. of records" FS $one FS $many; next} $one != "" {a[$one]++; b[$one][$many]++} END {for (i in b) {for (j in b[i]) {if (a[i]>b[i][j]) print b[i][j] FS i FS j} print RS}}' "$1" | cat -s | sed 's/^$/---------------/;s/\t/ | /g'
}
In the screenshot below, one2many finds multiple values of scientificNameAuthorship (field 49) for the same scientificName (field 9) in "CNS":

Notice that one2many has also found that authorship entries are missing for three of the scientificName entries. There can be good reasons why some entries might be missing from a Darwin Core field, but in this case those gaps shouldn't exist, because they've been filled for the same scientificName in other records. These gaps are missing-but-expected entries.
one2many does not tell you which of the multiple entries for one unique entry is the correct one. It only tells you that there's a data problem that needs fixing, even if the entries all looked OK when you checked them field by field.
Below are some of the field pairs worth checking with one2many:
unique value | multiple values?
scientificName | scientificNameAuthorship
scientificName | acceptedNameUsage
scientificName | taxonRank
scientificName | vernacularName
phylum | kingdom
class | phylum
order | class
family | order
subfamily | family
genus | family
country | continent
stateProvince | country
locality | country
locality | stateProvince
locality | county
locality | municipality
verbatimEventDate | eventDate
verbatimTaxonRank | taxonRank
verbatimLatitude | decimalLatitude
verbatimLongitude | decimalLongitude
scientificName | scientificNameID
scientificName | taxonID
recordedBy | recordedByID
nameAccordingTo | nameAccordingToID
one2many triplets
one2manyLL is the three-field version of one2many. In other words, it looks for multiple values associated with a pair of unique values in the same record. I often use this function for checking spatial fields when coordinates are provided. The unique two fields are decimalLatitude and decimalLongitude, and these are checked for multiple or missing values in a third field, such as country, as shown in the screenshot below with the table "CNS" (decLat. is field 19, decLon. 20 and country 27).

Some of the other fields against which coordinates might be checked are county, habitat, higherGeography, island, islandGroup, locationID, municipality, stateProvince and waterBody.
A reminder: this function only finds inconsistencies. It won't tell you if (for example) 57.85 12.033333 is in Sweden or in Switzerland. That particular coordinate check is one that GBIF does, and GBIF flags the errors with "coordinate country mismatch". You can do fine-scale coordinate checks one-by-one in an online mapper such as Google Maps, or bulk-wise in a GIS program.
A weakness of one2manyLL for coordinate checks is that it's based on exact string matching, so that the latitude "-35.0526" will be tested as different from the latitude "-35.052613", although these are the same for practical purposes.
A workaround is to apply the rounder function. This uses AWK to build a new, temporary table in which the last two fields have the coordinates rounded to four decimal places, and any records without lat/lons are excluded:
rounder() {
awk -F"\t" -v c1="$2" -v c2="$3" 'NR==1 {print $0"\tRoundedLat\tRoundedLon"} NR>1 {$(NF+1)=sprintf("%0.4f",$c1); $(NF+1)=sprintf("%0.4f",$c2); if ($NF != "0.0000") print}' OFS="\t" "$1" > "$1_rounded"
}
Use rounder like this:
rounder [filename] [latitude field] [longitude field]
In the screenshot below, rounder works on the 3-field file "demo", which only has an ID field (field 1), a latitude field (2) and a longitude field (3). The function builds the new table "demo_rounded":

A one2manyLL test on this new, temporary file (based on the new, rounded coordinate fields) shows a possible anomaly:

The choice of four decimal places for rounder is probably optimal. With three places and two places you might get agreements where there really aren't any, because you rounded too much, but five places might be too many. In the "demo" case, there would be no result because "35.05260" would not match "35.05261". There may also be occasional rounding issues with the way AWK does its rounding. This isn't a perfect test procedure, but it should work for the majority of Darwin Core datasets. In data tables where the event coordinates have the same number of decimal places, rounder isn't needed.
An interesting use for one2manyLL is to look for impossible or unlikely collecting events where the same collector (recordedBy) on the same day (eventDate) was apparently in two different places. A test with "CNS" turned up some anomalies worth further checking, as shown below with non-anomalies deleted. The checking would consider that one or more of three fields had incorrect entries: recordedBy (field 7), eventDate (33) or countryCode (38).
$ one2manyLL CNS 7 33 38
No. of records | recordedBy,eventDate | countryCode
---------------
6 | Stephens, S.E.,1943-03-21 | AU
1 | Stephens, S.E.,1943-03-21 | PG
---------------
1 | Edwards, K.,1998-03-19 | AU
1 | Edwards, K.,1998-03-19 | NZ
---------------
2 | Kilgour, C.D.,2011-04-29 | AU
1 | Kilgour, C.D.,2011-04-29 | MY
---------------
6 | Waterhouse, B.M.,1997-08-01 | AU
1 | Waterhouse, B.M.,1997-08-01 | ID
---------------
Pairs and triplets
The fldpair function shows the counts of records in pairs of fields.
fldpair() {
awk -F"\t" -v one="$2" -v two="$3" 'NR==1 {x=$one; y=$two; next} ($one!="") && ($two!="") {both++} ($one!="") && ($two=="") {oneonly++} ($one=="") && ($two!="") {twoonly++} ($one=="") && ($two=="") {neither++} END {print "neither\t"neither"\n"x" only\t"oneonly"\n"y" only\t"twoonly"\nboth\t"both}' "$1" | sed 's/\t$/\t0/' | column -t -s $'\t'
}
fldpair is useful for quickly checking whether logically paired fields are filled. For example, both decimalLatitude and decimalLongitude should either be both filled or both blank, and this is true for "CNS":

The use of elevation fields, however, is inconsistent in "CNS" (see screenshot below). It would be better to use only one of the fields, or preferably both.

There are many possible field pairs you can check with fldpair, although the use of this function overlaps with use of one2many.
The 3-field version of fldpair is fldtriplet:
fldtriplet() {
awk -F"\t" -v one="$2" -v two="$3" -v three="$4" \
'NR==1 {x=$one; y=$two; z=$three; next} \
($one != "") && ($two != "") && ($three != "") {allthree++} \
($one != "") && ($two == "") && ($three == "") {oneonly++} \
($one == "") && ($two != "") && ($three == "") {twoonly++} \
($one == "") && ($two == "") && ($three != "") {threeonly++} \
($one != "") && ($two != "") && ($three == "") {onetwoonly++} \
($one == "") && ($two != "") && ($three != "") {twothreeonly++} \
($one != "") && ($two == "") && ($three != "") {onethreeonly++} \
($one == "") && ($two == "") && ($three == "") {none++} \
END {print \
"none\t" none "\n" \
x " only\t" oneonly "\n" \
y " only\t" twoonly "\n" \
z " only\t"threeonly "\n" \
x " + " y " only\t" onetwoonly "\n" \
x " + " z " only\t" onethreeonly "\n" \
y " + " z " only\t" twothreeonly "\nall three\t" allthree}' "$1" \
| sed 's/\t$/\t0/' | column -t -s $'\t'
}
fldtriplet is particularly handy for looking at decimalLatitude, decimalLongitude and geodeticDatum as a group. In the screenshot below, fldtriplet checks "oct" and finds there is latitude but no longitude in five records, and there is a datum without any coordinates in 40 records:

Between name fields
In datasets with species-level scientific names and their components genus and specificEpithet, a useful check is to see whether scientificName agrees with those components. This command will do that check in an occurrences table (occurrence.txt):
awk -F"\t" 'NR>1 {split($[scientificName],a," "); if (a[1]!=$[genus] || a[2]!=$[specificEpithet]) print $[occurrenceID] FS $[scientificName] FS $[genus] FS $[specificEpithet]}' occurrence.txt
The command splits scientificName into its components with AWK's "split" function and it relies on there being one or more plain spaces between the genus and species epithets. The command will fail if (as is too often the case!) the space is an invisible no-break space (NBSP), so NBSPs need to be converted to plain spaces before doing this between-field checking.
Because this command is a little long, it pays to store it in a "generic" form in a text file, then paste the "generic" into the terminal and edit it before launching. My "generic" command looks like this:
awk -F"\t" 'NR>1 {split($N,a," "); if (a[1]!=$G || a[2]!=$S) print $O FS $N FS $G FS $S}' file | wc -l
where N = number of scientificName field, G = number of genus field, S = number of specificEpithet field and O = number of occurrenceID or other unique identifier field. The command ends with wc -l so I can see how many records have disagreements; I may need to page the results through less to see them all. In the table "occurrence.txt" shown below, the command found six disagreements.

Note the "." in specificEpithet in that last record. That formatting error should have been fixed during the within-field stage of the data checking!
The checking command would be even longer and more complicated if it also looked at infraspecificEpithet, because the infraspecific epithet might be separated from the species name in scientificName by a blank, "subsp.", "var." etc. My practice is to check all scientificName entries by taxon rank, like this:
awk -F"\t" '$[taxonRank]=="[taxonRank entry]" {print $[scientificName]}' [filename] | sort | uniqc | less
and then check the entries by eye (but see below). For infraspecies I include the infraspecificEpithet field in the output:
awk -F"\t" '$[taxonRank]=="[infraspecificEpithet entry]" {print $[scientificName] FS $[infraspecificEpithet]}' [filename] | sort | uniqc | less
When the list of names to be scanned is long and hard to scan by eye, it can help to do a regular expressions check. For example, a dataset might have hundreds of names with taxon rank "species", but some infraspecies may have been given the "species" rank by mistake.
awk -F"\t" '$[taxonRank]=="species" {print $[scientificName]}' [filename] | sort | uniq | grep -E "^[A-Z][a-z]+ [a-z]+ [a-z]+"
Here the regular expression ^[A-Z][a-z]+ [a-z]+ [a-z]+ after grep will find "Genus species subspecies", "Genus species subsp. something", "Genus species var. something", etc. If scientificName includes authorship, the regex will give a false positive result with a name such as "Genus species van Loon, 1823".
Another between-field check that's hard to do by eye is to compare scientificName with scientificNameAuthorship when authorship is repeated as part of the scientificName entry. I use the following function, which takes four arguments: filename, number of the scientificName field, number of the scientificNameAuthorship field and number of the field holding the record's unique ID:
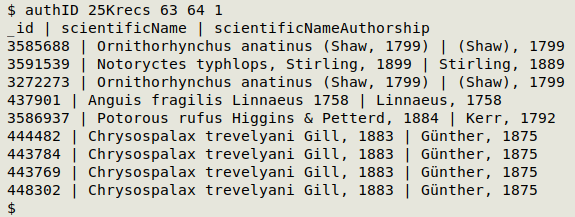
authID() { awk -F"\t" -v sciN="$2" -v sciNA="$3" -v ID="$4" 'NR==1 {print $ID FS $sciN FS $sciNA; next} $sciN != "" && $sciNA != "" && index($sciN,$sciNA)==0 {print $ID FS $sciN FS $sciNA}' "$1" | barsep; }
The output is a list with unique ID code, scientificName and scientificNameAuthorship for records in which the two authorship strings are not exactly the same. Note that the list will also include records where the scientific name does not include any authorship; this is usually the case with taxa above genus level. Here is the command at work on the Darwin Core table "25Krecs" (25000 records), where the ID field is 1, scientificName is field 63 and scientificNameAuthorship is field 64:
Between date fields
eventDate cannot logically be later than dateIdentified, measurementDeterminedDate or modified. eventDate could be later than georeferencedDate if the observation or collection site was selected and georeferenced in advance.
The job of checking this logic is easy if all the dates are in YYYY-MM-DD format. This is because a command-line sort of ISO 8601 dates is automatically chronological, and ">" is interpreted by AWK as "appears later in a sort of ISO 8601 date strings". To check if eventDate is later than dateIdentified in an occurrence.txt table with YYYY-MM-DD dates:
awk -F"\t" 'NR>1 && $[eventDate] > $[dateIdentified] {print $[eventID] FS $[eventDate] FS $[dateIdentified]}' occurrence.txt
In "demo1", below, this check has found 76 records with eventDate (field 2) later than dateIdentified (field 3; printed with occurrenceID, field 1):

The check is more complicated if not all the dates are in YYYY-MM-DD format. For example, "2023-06-25" in eventDate is "later" than "2023-06" in dateIdentified by the sorting rules, but there isn't an error here, because the day of identification has not been specified.
In an occurrence.txt file with a mix of YYYY, YYYY-MM and YYYY-MM-DD dates, we can check for records where eventDate YYYY is later than dateIdentified YYYY, YYYY-MM is later than YYYY-MM, and/or YYYY-MM-DD is later than YYYY-MM-DD with the 2datesID function:
2datesID() {
awk -F"\t" -v earlier="$2" -v later="$3" -v idfld="$4" 'NR==1 {print $idfld"\t"$earlier"\t"$later} NR>1 && $earlier != "" && $later != "" {split($earlier,a,"-"); split($later,b,"-"); if ((a[1] > b[1]) || (a[2] && b[2] && a[1]"-"a[2] > b[1]"-"b[2]) || (a[3] && b[3] && a[1]"-"a[2]"-"a[3] > b[1]"-"b[2]"-"b[3])) print $idfld FS $earlier FS $later}' "$1" | barsep'
}
2datesID takes four arguments: filename, earlier date field, later date field, unique ID field. In the screenshot below, 2datesID checks "demo2" and finds 313 records where eventDate (field 2) is later than dateIdentified (3), and also prints the unique ID field occurrenceID (1):

Note: The toptail function used above prints the first and last 10 items in a list. The code is
toptail() {
sed -u '10q'; echo "---"; tail
}
Many Darwin Core tables include year, month and day fields where eventDate is YYYY, YYYY-MM and/or YYYY-MM-DD. To check for disagreement between eventDate and these component date items in (for example) an event.txt file:
awk -F"\t" 'NR>1 {split($[eventDate],a,"-"); if (a[1]!=$[year] || a[2]!= $[month] || a[3]!=$[day]) print $[eventID] FS $[eventDate] FS $[year] FS $[month] FS $[day]}' event.txt
In "demo3" (screenshot below)), occurrenceID is field 3, eventDate is field 7, year is field 8, month is field 9 and day is field 10. There are 24 records with disagreements.

Please note that the date checks shown above are for single dates. Checking interval dates (e.g. YYYY-MM-DD/YYYY-MM-DD) can also be done with AWK after splitting the interval date on the "/" character. This can be tricky to do because it depends on the variety of date formats in the dataset.
Between-field miscellany
minimumElevationInMeters and maximumElevationInMeters (or their depth equivalents)
The entries in these two fields are sometimes swapped. To check for minimum greater than maximum, a first condition should be that both fields have entries, i.e. their entries are not the empty string (""):
awk -F"\t" 'NR>1 && $[minEIM] != "" && $[maxEIM] != "" && $[minEIM] > $[maxEIM] {print $[ID field] FS $[minEIM] FS $[maxEIM]}' [filename]
individualCount (or organismQuantity) and occurrenceStatus
Valid entries for occurrenceStatus are "present" and "absent". If there is a non-zero entry in individualCount (or organismQuantity), the entry in occurrenceStatus should be "present", not "absent" or blank:
awk -F"\t" 'NR>1 && $[individualCount] != 0 && $[individualCount] != "" && $[occurrenceStatus] != "present" {print $[ID field] FS $[occurrenceStatus] FS $[individualCount]}' occurrence.txt
basisOfRecord and catalogNumber for "PreservedSpecimen"
In museum or herbarium datasets where basisOfRecord is "PreservedSpecimen", not all the records may have an entry in catalogNumber. This might be because the museum sample lot or herbarium sheet is not yet registered, or because the registration code was left out by mistake. To find these records:
awk -F"\t" '$[basisOfRecord] ~ /Pres/ && $[catalogNumber] == "" {print $[occurrenceID]}' occurrence.txt
scientificName and establishmentMeans
In a faunal or floral study of a single area, such as a nature reserve, the entry in establishmentMeans (e.g. "native", "introduced", "vagrant", "uncertain" etc) would be expected to have the same value in all records for a single taxon. Check with a one2many test:
one2many [filename] [scientificName] [establishmentMeans]
eventID and parentEventID
When eventID codes are built from parentEventID the agreement between the two can fail, and it can be very hard to spot disagreements among hundreds of code pairs. There is no general method for finding disagreements, because the method has to be based on the particular code structures. However, an AWK command will almost always work:

eventDate and georeferenceSources
This is an odd one, but I did look at a dataset in which there were numerous records dated well before the 1990s in which georeferenceSources had "GPS". To find the years-with-GPS I did:
grep GPS [filename] | cut -f[eventDate] | cut -d"-" -f1 | sort | uniq
When I emailed the data compiler, he told me that the records were old ones, but that the compiler and colleagues had re-visited the old localities, and the coordinates in those records were based on recent GPS readings.
basisOfRecord and references
In a herbarium dataset I audited, almost all basisOfRecord entries (field 6) were "PreservedSpecimen", but 280 records were "HumanObservation". A references field (field 92) contained URLs to herbarium sheet images and some blank entries. How many of the "HumanObservation" records had sheet image URLs, and were therefore actually "PreservedSpecimen" records?
eventDate and associatedReferences publication date
In an occurrence.txt table with MaterialCitation sources for occurrence records, I found event dates that were later than the publication date of the reference, which is not possible. All the citations in associatedReferences (field 6) had years in the form "(YYYY)" or "(YYYY[a,b etc])". occurrenceID was field 5 and eventDate was field 7. I used AWK to find the anomalies:
awk -F"\t" '$6 ~ /[0-9]{4}/ && $7 != "" {split($6,a,/[0-9]{4}/,s); split($7,b,"-"); if (s[1] < b[1]) print $5 FS $6 FS $7}' occurrence.txt | barsep | sed G

To explain: AWK first selects those records in which associatedReferences has a match for four digits in a row (the year number) and eventDate is not blank. It then splits the reference entry using the 4-digit number as a separator, and puts the separator(s) in an array "s". Next, AWK splits the ISO 8601 event date using "-" as separator and puts the components in an array "b". Next, AWK looks for records in which the first separator in the reference split (the reference year) is less than (before) the event year. It prints occurrenceID, associatedReferences and eventDate from these records. sed G spaces the records. This is a good example of the "ad hoc" checking for which AWK is so useful.
